Structural Biochemistry/Volume 7
Carbohydrates
[edit | edit source]Classification
[edit | edit source]Monosaccharides are the simplest form of carbohydrates and may be subcategorized as aldoses or ketoses. The sugar is an aldose if it contains an aldehyde functional group. A ketose signifies that the sugar contains a ketone functional group. Monosaccharides may be further classified based on the number of carbon atoms in the backbone, which can be designated with the prefixes tri-(3), tetr-(4), pent-(5), hex-(6), hept-(7), etc. in the name of the sugar.
Monosaccharides are often represented by a Fischer Projection, a shorthand notation particularly useful for showing stereochemistry in straight chained organic compounds. The L and D confirmations represent the absolute configuration of the asymmetric carbon farthest away from the ketone or aldehyde group on the monosaccharide. On the Fischer projection, if the farthest hydroxyl(-OH) group is on the right, then it is classified as D sugar, if the hydroxyl group is on the left, then it is a L sugar.
Enantiomers, Diastereoisomers(anomerism), and Epimers
[edit | edit source]

Due to the fact that carbohydrates contain multiple stereocenters, many isomers are possible including enantiomers, diastereoisomers, and epimers.
Two carbohydrates are said to be enantiomers if they are nonsuperimposable mirror images of one another. An example of an enantiomer is the D and L isomers of glucose, as shown by the figure to the right.
A second type of isomer seen in carbohydrates are diastereoisomers. Carbohydrates are classified as diastereomers if their chiral carbons are connected to the exactly the same substrates but connected at differing configurations (R or S). Unlike an enantiomer, diastereomers are NOT object and mirror image. An example of two carbohydrates that are diastereoisomers are D-Glucose and D-Altrose as seen in the figure to the left.
Lastly, another type of isomer that carhbohydrates that can take on are epimers. Epimers are two diastereomers that differ only at one stereocenter.[1] As shown in the figure below, D-Glucose and D-Mannose are an example of an epimer.

Simple Aldoses
[edit | edit source]An Aldose contains an aldehyde with two or more hydroxyl groups attached; one of the hydroxyl groups is at end opposite to the aldehyde. An Aldose is a type of monosaccharide, which is a chiral molecule that plays a key role in the development of nucleic acids. The two simplest forms of Aldoses are L- and D-Glyceraldehydes, which are three-carbon structures that each contain one aldehyde and two hydroxyl groups. The L and D symbols apply to the two different configurations of the asymmetric carbon farthest from the aldehyde group. Aldoses can have three or more carbons. Aldoses are distinguishable by the carbonyl(C=O) group located at the end of the carbon chain, which differs from ketose, which has the carbonyl group in the middle of the carbon chain. Furthermore, for example, the simplest sugar with three carbons - glyceraldehyde (containing an aldehyde group), and sugar with seven carbons - L-glycero-D-manno-heptose, can be found in this category. Each sugar has n = C - 2 numbers chiral center(s), where C is the number of carbons. We can also use the formula 2n to calculate the maximum number of stereoisomers that are possible to exist in a molecule. Again, n is the number of stereocenter(s). For example, aldotriose has three carbons(C), one stereocenter(n) - it has two stereoisomers. Following the same calculation, we know that aldotetroses have four stereoisomers, aldopentoses have eight stereoisomers, and aldohexoses have sixteen stereoisomers. We usually focus on the D sugars since they are more frequently in natural existence, while the L sugars, diastereomer of the D sugars, are less common. See Chirality for the naming using D/L system.
List of common aldoses
[edit | edit source]
Triose
[edit | edit source]Trioses are monosacchrides with three carbon atoms; aldotrioses have an aldehyde functional group at carbon number one. A common aldotrisose is glyceraldehyde. They have a single asymmetric carbon atom: D- and L-glyceraldehyde are enantiomers of one another. Glyceraldehydes are one of the smallest monosaccharides.
Tetroses
[edit | edit source]
Tetroses are monosaccharides with four carbon atoms. An aldotetrose has an aldehyde functional group at carbon number one.
The two common types of aldotetroses are D-Erythose and D-Threose. The D configuration is more favor. Since D-Erythose and D-Threose are not mirror image of one another, they are diasteroisomers of one another. They have a different configuration at the second carbon. They have two asymmetric carbons and four steroisomers.
Pentoses
[edit | edit source]A Pentose is a type of monosaccharide which has a backbone of five carbon atoms. At carbon position 1, there is an aldehyde functional group attached which gives it their aldose nature.
Common types of 5-Carbon Aldoses include Ribose, Arabinose, Eibose, Lyxose, and Xylose

Hexoses
[edit | edit source]

A hexose is a monosaccharide with six carbons, but more specifically, an aldohexose is a hexose with an aldehyde functional group at carbon number one.
Some common aldohexoses are Allose, Altrose, Glucose, Mannose, Gulose, Idose, Galactose, Talose.

A trick to remember names and structures of hexose in D-aldose configuration:
1. Use a sentence "All altruists gladly make gum in gallon tanks" to write down in order allose, altrose, glucose, mannose, gulose, idose, galactose, talose.
2. Draw Fisher projections for those hexoses with C-1 is -CHO group, C-6 is _CH2OH
3. Now we add the OH group, start from C-5 and from allose to Talose (from left to right):
a. at C-5 : all -OH will be attached on the right
b. at C-4: -OH will be attached on the right for the first 4 hexose (allose to mannose), and -OH will be attached on the left for the last 4 hexose (Gulose to Talose)
c. at C-3: 2 right, 2 left, 2 right, 2 left
d. at C-2: right, left, right, left, and so on.
Cyclic Monosaccharides
[edit | edit source]
Alodopentoses and aldohexoses can exist in three different forms: the open chain as appears in the Fisher projection, the two cyclic forms of alpha(α) sugar and beta(β) sugar. Ring formation tends to be energetically more stable than open chains. Pentoses often cyclize into a ring form structure called Furanose whereas hexoses form cyclic sugars called pyranoses. The two different forms of cyclic sugars, alpha and beta, are referred to as anomers. For example, in D-glucose, the hydroxy group on carbon 5 attacks the carbonyl carbon forming a six membered ring with the carbon that was attached, being known as the anomeric carbon. The resulting hemiacetal sugar is known as a pyranose. α-D-glucose is formed if the newly formed hydroxyl group is pointed in an opposite direction to the CH2OH group in Haworth projection, and β-D-glucose is formed if the hydroxyl group is pointed in the same direction as the CH2OH group. The majority, about 66% of D-glucose exist in β form because when the molecule is in chair conformation, all the bulky hydroxyl groups will be placed in equatorial position - which have lesser steric hindrance between the bulky groups. Thus, β-D-glucose is more stable than α-D-glucose that occupied typically 33% of D-glucose molecules, whereas the remaining 1% is in the open-chain form.
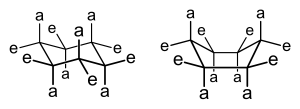
There are two different conformations a pyranose and furanose ring can take: chair and boat form. In the chair form there are two different orientations: equatorial and axial positions. In the axial position, the substituents will form bonds that are perpendicular to the plane of the ring. However, these bonds may often form a steric hindrance due to crowding of the substituents. If two substituents are near and pointing within the same direction, then there will be a steric hindrance. In contrast, the equatorial position will have its substituents to form bonds that are parallel to the plane of the ring. This formation produces less crowding and is the most preferred form for the chair conformation. Lastly, the boat form is unlikely, due to the fact that there is crowding and steric hindrance.
Haworth Projection
[edit | edit source]A Haworth projection is a simple way to show cyclic sugars and their glycosidic linkages. It consists of the ring on a horizontal plane but ignores the chair and boat forms so that the ring is flat. If drawn from a Fischer projection with the carbonyl on top, the groups on the right side become the groups on the bottom of the ring and the groups on the left become the groups on the top. The carbon at the very bottom of the Fischer projection is placed on top of the ring by default (if it is not a part of the ring itself). An α-linkage occurs when the hydroxyl of the hemiacetal is on the bottom of the right and a β-linkage occurs when the hydroxyl is on top.
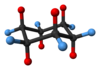
If drawing from a chair form, all groups on top of the chair become the groups on top of the Haworth projection ring. The easiest way to find all these groups is to choose an axial position that is above the ring and then go around the ring, alternating axial and equatorial positions. In the picture on the right, all the groups attached to red bonds would go on top of the ring the Haworth projection and all the groups attached to the blue bonds would go below.
Simple Ketoses
[edit | edit source]A ketose is a sugar that has a ketone group in each of its molecule. Dihydroxyacetone, for example, has 3 carbon atoms in its backbone - it is the simplest ketose among this category. It is also the only optically inactive ketose. In comparison to other aldoses, ketones will have one less chiral carbon than aldoses even though they share the same number of carbon atoms. Thus, when forming a ring, the ketone at the second carbon will be utilized to form a ring.
Similar to aldoses, furanose rings can take up a different conformation than a ring. The other conformation is called the envelope form: C3-endo and C2-endo. Both forms resembles an envelope.

List of common ketoses
[edit | edit source]Triose: A triose contain 3 carbon, and ketotriose contains a ketone functional group. A ketotriose has no chiral center and one stereoisomers. An example of ketotriose is Dihydroxyacetone. Dihydroxyacetone has many uses, and it is non-toxic. Many creams had Dihydroyactenone as an active ingredient. Dihydroxyacteone is also known as DHA. It is also use for suntanning. (Chemical and Engineering News)

Tetrose: Erythrulose A Tetrose is a monosaccharide that contains 4 carbon atoms. A Keto-tetrose is a tetrose that has a ketone functional group attached to Carbon 2 of the straight chain. A ketotetrose has 2 stereoisomers because it has one chiral center. An example of a ketotetrose is Erythrulose. Erythrulose has the chemical formula of C4H8O4. It is often used in self-tanning products.

Pentoses: Ribulose, Xylulose A Pentose is a general term to define a monosaccharide containing five carbons. When there is the prefix "keto" in front of the pentose, it means that in five carbon containing sugar, there is a ketone functional group attached to the structure. A ketopentose has a total of four stereoisomers. An example of a ketopentose is Ribulose. The structure of Ribulose has a ketone functional group attached to C-2 of the straight chain figure. The diastereomer of D-Ribulose is D-Xylulose.
Hexoses: A Hexoses contains 6 carbons. A hexoses containing a ketone functional group is called ketohexose. Ketohexose has 3 chiral centers and 8 different stereoisomers. Examples of ketohexose are Fructose, Psicose, Sorbose, Tagatose. Fructose can react with hydroxyl group to form a hemiketal group, and it can formed pyranose or furanose depending on whether the C-2 keto group reacts with the C-6 or C-5 hydroxyl group. D-Fructose is the most common ketohexose.
Ketoses in Reactions
[edit | edit source]Transketolase Reaction
[edit | edit source]The Transketolase reaction is very similar to the Transaldolase reaction. However, the Transketolase is different because it transfers a two carbon unit instead of Transaldolase's three carbon unit. Thiamine pyrophospate (TPP) ionizes so that it has a carbanion which is a negatively charged carbon. The importance of carbanion is that they can attack carbonyls, so that carbons are added in a sense to the nucleophile. TPP attacks a ketose substrate where it than releases the aldose product to yield an activated glycoaldehyde unit. An activated glycoaldehyde unit is an electron sink because of a positively charged nitrogen atom where a carbonyl of an aldose product is attacked and then separated after some electron movement. The importance of the transketolase reaction is that it is the mechanism that the enzyme TPP uses to change a ketose substrate to a ketose product that has a different group attached to it.
Transaldolase Reaction
[edit | edit source]The transaldolase reaction involves the transfer or a three carbon dihydroxyacetone unit from a ketose donor to an aldose acceptor. Unlike the transketolase reaction, transaldolase does not contain a prosthetic group; instead the reactions begins with a Schiff base formed between the carbonyl group of the ketose substrate and the amino group of a lysine residue at the active site of the enzyme. Next the Schiff base is protonated and the bond between C-3 and C-4 break which releases the aldose product. The leftover negative charge on the Schiff-base carbanion is stabilized by resonance while the positive charge on the nitrogen atom of the protonated Schiff base acts as the electron sink. The Schiff-base remains stable until a suitable aldose becomes bound which allows the dihydroxyacetone to react with the carbonyl group of the aldose and the ketose product is released from the lysine side chain via hydrolysis of the Schiff-base.
Transaldolase is a target of autoimmunity in patients with multiple sclerosis which is the selective destruction of oligodendrocytes that selectively expresses transaldolase in the brain.
Ketose in the Calvin Cycle
[edit | edit source]The Calvin cycle, or dark reactions, is one of the light-independent reactions. In the third phase of the this reaction, a five-carbon sugar is constructed from six-carbon and three-carbon sugars. A transketolase and an aldolase are the major factors in the rearrangement. The transketolase, which is in the pentose phosphate pathway, requires a coenzyme, thiamine pyrophosphate (TPP), to transfer a two-carbon unit from a ketose to an aldose. Whereas the transaldolase transfers a three-carbon unit from a ketose to an aldose.
In summary, transketolase first converts a six-carbon sugar and a three-carbon sugar into a four-carbon sugar and a five-carbon sugar. Then, aldolase combines the four-carbon product and a three-carbon sugar to form the seven-carbon sugar. This seven-carbon sugar then finally reacts with another three-carbon sugar to form two additional five-carbon sugars.
Energy for Organic Organisms
[edit | edit source]Glucose (C6H12O6) is one of the main products of the photosynthetic process by plants that initiates the cellular respiration process that produces ATP (adenosine triphosphate), the basic energy currency for prokaryotes and eukaryotes. Glucose is also involved in the energy-harvesting process of glycolysis, which converts glucose into pyruvate and yields a much lesser amount of ATP than is produced by the electron transport chain within cellular respiration. Glucose is an essential source of energy for the body.
Modified monosaccharides
[edit | edit source]
One example of modified monosaccharides are the phosphorylated sugars. An important phosphorylated sugar is glucose 6-phosphate, which is a glucose phosphorylated on carbon 6. The significance of this molecule is that it provides energy in certain metabolic pathways, and it can be converted and stored as glycogen when blood glucose levels are high. If blood glucose levels are low, glucose 6-phosphate can be converted back into glucose to enter the bloodstream once again. A unique property of glucose 6-phosphate is that once glucose is phosphorylated, the sugar possesses a negative charge. This prevents the molecule from leaving the lipid-bilayer membranes. This allows the cell to easily access the modified sugar to provide energy for metabolic pathways such as glycolysis, or convert it to glycogen as storage.
Importance of Carbohydrates in Nature
[edit | edit source]The biological significance of carbohydrates is unquestionable in the natural world with its essential roles in providing metabolic energy. Carbohydrates not only serve roles in energy, but also storage and plant cell wall structure; however carbohydrates are also involved in a variety of biological processes including the immune response, cell–cell interactions, fertilization, viral infection, and drug efficacy, among others. In recent years, researchers are discovering and understanding new sugar moieties that may have important ramifications for the development of new therapeutics. For example, the dideoxysugar and trideoxysugar moieties that are synthesized by a wide range of bacteria, fungi, and plants are representation of a captivating class of carbohydrates. They are found on the lipopolysaccharides, on the S-layers of some Gram-positive and Gram-negative bacteria, on extracellular polysaccharides, and on antibiotic, antifungal, anthelmintic, and antitumor agents. These diverse complex carbohydrates are derived from simple monosaccharides such as glucose-6-phosphate or fructose-6-phosphate that goes through numerous of enzymatic reactions including acetylations, aminations, dehydrations, epimerizations, reductions, and methylations. The bacterial N-acetyltransferases and the PLP-dependent aminotransferases are enzymes for the biosynthesis of unusual dideoxysugars and trideoxysugars. With the understanding of the structures and the functions of these enzymes that are required for the biosynthesis of the sugars, this makes it possible to redesign new drugs that will only benefit humans because these sugar moieties are only synthesized in bacteria, fungi and plants.
References
[edit | edit source]- ↑ Organic Chemistry Structure and Function, Sixth Edition.
Berg, Jeremy M. John L. Tymoczko. Lubert Stryer. Biochemistry Sixth Edition. New York: W.H. Freeman, and Company 2007.
Chemical and Engineering News. http://pubs.acs.org/cen/whatstuff/stuff/7824scit2.html/ carbohydrates are synthesize by energy process that is photosynthesis which is take place in green plants in presence of co2+h2o .splar energy in green pigments that is chlorophylland this process is called photosynthesis
Nomenclature
[edit | edit source]Origin
[edit | edit source]
Glyceraldehyde serves as the basis in naming monosaccharides since it is the simplest monosaccharide, having only one asymmetric carbon. (+)-Glyceraldehyde was arbitrarily named the D-enantiomer (the hydroxy group is on the right when drawn as a Fischer Projection). Proof that the structure matched the optical rotation was not obtained until many years later.
(+) and (-) enantiomers
[edit | edit source](+) enantiomers rotate plane-polarized light clockwise (also called dextrorotary, abbreviated d), while (-) enantiomers rotate it counter-clockwise (levorotary, or l). This must be determined empirically.
D and L enantiomers
[edit | edit source]D and L enantiomers refer to the configurational stereochemistry of the molecule. L isomers have the hydroxy group attached to the left side of the asymmetric carbon furthest from the carbonyl, while D isomers have the hydroxy group on the right side. Many naturally occurring sugars are D isomers, although exceptions include arabinose and rhamnose. This system of nomenclature is NOT necessarily the same as optical rotation (D and L are not the same as d and l). In other words, D and L configurations do not fully designate absolute stereochemistry, rather they are determined on the basis of the anomeric carbon center and how its orientation compares to glyceraldehyde, the most basic and simple chiral sugar molecule. It is mostly in aldoses and is not present in ketoses.
R and S enantiomers
[edit | edit source]
Like naming sugars based on D and L, the asymmetric carbon furthest from the carbonyl is the one that determines the name.
All D sugars are R isomers because they all have the hydroxyl group attached to the right of the last asymmetric carbon. By the Cahn-Ingold-Prelog rules for naming stereochemistry, the hydroxyl group will always be priority 1, the carbon of the primary alcohol (the terminal carbon) will always be priority 3, the rest of the carbon chain will be priority 2, leaving hydrogen as priority 4 (as shown below). With the hydroxy on the right, the carbon of interest will always be an R isomer.
The example on the right shows D-Glucose with priorities of each substituent numbered. When rotated to view down the C-H bond, the priorities decrease in a clockwise fashion, hence that stereocenter is designated R. However, for the enantiomer of D-glucose, the priorities decrease in a counterclockwise fashion indicating that the stereocenter is designated S.
Fischer Stereochemistry Proof
[edit | edit source]Herman Emil Fischer presented the stereochemical configuration relationship in sugar through a series of experiments with ribose. At the time when this experiment was conducted, all they had was optical rotation to determine stereochemistry. Optical rotation assigns (+) for one enantiomer and (-) for the opposite one. However there were no direct correlations with (+/-) and (R/S) for all chiral sugars. For example, for a particular sugar, the R form may be (+) and the S form (-), but in another sugar, the R may be (-) and the S form (+). Fischer was able to manipulate a series of reactions to assign stereochemistry among sugars. At first he just assumed the penultimate position of the experimental arabinose was in R-configuration. He had a 50/50 chance of picking the correct conformation and if, in the future, the experimental arabinose turn out to be in L form, all his data is still relatively correct, just inverted. Luckily, the arabinose was later proved to be in D-conformation.
Under the Kiliani-Fischer synthesis condition, arabinose will produce two epimeric sugars, mannose and glucose. Although it remained unknown which one was glucose and which one was mannose.
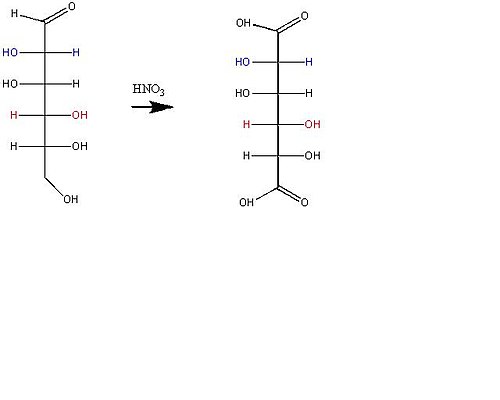
By adding the HNO¬¬3 to arabinose, arabinose will be oxidized into an optically active aldaric acid. Out of the four possible aldaric acid derivatives from a set R penultimate configuration, two were eliminated because they were not optically active. The two remaining candidates’ C2 have the same S stereocenter configuration.
Possible aldaric acid
Next, mannose and glucose were oxidized by HNO3. Mannaric acid and glucaric acid were also optically active. With only one unknown stereocenter, there are two possible forms of aldaric acid for each sugar. Out of the four total predictions of glucose and mannose, one of the aldaric acids is meson and therefore cannot be either Mannaric or glucaric acid. Mannaric acid and glucaric acid should have the same stereocenters except for the inverted C2 stereocenter. When one of the models below was rejected, the other model whose C4 is in an S configuration was also rejected. Below, the two circled aldaric acids are mannaric acid and glucaric acid.
The last part of the Fischer proof was to figure out which one is actually glucose. The last clue to Fischer’s proof was that, while glucaric acid can be derived from two sugars, mannaric acid can only be derived from the oxidation of mannose, because mannaric acid is rotationally symmetrical.
Glucaric acid:
Mannaric Acid:
Haworth projection
[edit | edit source]Haworth projection is used to present cyclic hemiacetals. These followings are steps to convert monosaccharides to cyclic hemiacetals:
- Choose the position for the oxygen
- The oxygen is in the back right-hand corner of the ring (for six-membered ring.)
- The oxygen is away from the viewer (for five-membered ring.)
- The left of the oxygen is C5 with the hydroxymethyl group drawn up.
- The right of the oxygen is C1 with hydroxyl group drawn up or down depending on the α or β structure.
- –OH groups on the right side of the Fischer projection is drawn down.
- –OH groups on the left side of the Fischer projection is drawn up.
D&L enantiomers
[edit | edit source]If these positions are switched, you will instead have the L (-) enantiomer of glyceraldehyde. For monosaccharides, D and L will be used as prefixes instead of R and S, respectively, in regards to stereochemistry. The stereochemistry of all other monosaccharides can be determined by comparing their Fischer projections to that of D-(+)-Glyceraldehyde. This can be done by examining the stereocenter in the monosaccharide closest to the terminal carbon (the highest-numbered stereocenter)and comparing its configuration to that of glygeraldehyde. That is, if the hydroxy group is on the right, it will be named D- and if the hydroxy group is on the left it will be named L-. It is important to note that for all monosaccharides other than glyceraldehyde, the labels D and L do not necessarily say anything about its optical rotation. For instance, D-Glucose and D-Gulose have both been assigned the stereochemical label D due to their highest-numbered stereocenter (the chiral center furthest from the carbonyl group) having a hydroxy group on the right in their Fischer projections despite Glucose having a positive (dextro-) optical rotation and Gulose having a negative (levo-) optical rotation.


Alpha vs Beta Anomers
[edit | edit source]Hexoses and pentoses can convert to cyclic pyranoses or furanoses. As these monosaccharides convert between their linear and cyclic formations, the hydroxyl group on the C5 or C6 carbon can attach on either side of the carbonyl of C1 (as shown in image above). If the hydroxyl group is pointed in the opposite direction of the CH2OH group, the ring is in its alpha form. However if it is pointed in the same direction, the ring is in its beta form.


Diastereomers and Epimers
[edit | edit source]Two non-identical monosaccharides are said to be diastereomers if they are of the same type (either both aldoses or both ketoses), have the same stereochemistry at their highest-numbered stereocenter, and have the same number of carbons (i.e. are both tetroses). This is because having the same stereochemistry at their highest-numbered asymmetric carbon ensures that the two non-identical monosachharides will not be mirror images of each other and are therefore not enantiomers. Two monosaccharides that are diastereomers that have differing stereochemistry at only 1 asymmetric carbon (this carbon cannot be the highest-numbered asymmetric carbon) are called epimers. For instance, (D-Glucose and D-Mannose) and (D-Glucose and D-Galactose) are both epimers and diastereomers.
Conformational Isomers
[edit | edit source]Hexoses and pentoses that have converted into pyranoses or furanoses take on either chair, boat, or envelope conformations due to the tetrahedral geometry of their carbons. Pyranose rings can form either chair or boat conformational isomers (conformers) while furanose rings take on the envelope (also called half-boat) conformation. Substituents on the carbons in the monosaccharides are now either in axial or equatorial positions. The favored conformational isomer will be that which is the least sterically hindered, often containing the majority of its bulkier substituents in equatorial positions, since substituents in axial positions on the same side of the ring create steric hindrance. The chair conformation of pyranose rings can also undergo a ring flip, which switches the orientation of substituents from axial to equatorial and vice-versa, to produce an additional conformational isomer. Chair conformation of six-membered rings is most favorable as it reduces steric interference between two carbon substituents. Boat and Envelope conformations do not exist, but are theorized to act as an intermediate structure existing briefly between a ring flip transition in which axial substituents become equatorial and vice versa.

Sources
[edit | edit source]Fischer stereochemistry proof: http://nebula2.deanza.edu:16080/~gray/pages/chem_12c.html image was done on the ChemArt program and Paint One important method of elucidating the structure of carbohydrates is using periodic acid degradation. Periodic acid (HIO4) is a reagent that cleaves the carbon-carbon bonds in a sugar through oxidation. Periodic acid attacks the vicinal diols in carbohydrates and oxidizes these groups to form carbonyl compounds. The mechanism of this reaction involves a cyclic periodate ester that reacts with two neighboring alcohol functional groups which are oxidized to carbonyl functional groups.
How is a sugar degraded through periodic acid degradation?
[edit | edit source]When a sugar is reacted with excess periodic acid, each carbon-carbon bond is broken, forming a characteristic composition of one-carbon compounds that can provide some information about the structure of that carbohydrate. A quick rule of thumb for this reaction is that for any given carbon atom in the carbohydrate, for each carbon-carbon bond that is broken, that carbon atom will be oxidized once. Therefore, each broken bond will be replaced with an OH group. If there is a any carbon with two OH groups, it will lose water and become a carbonyl group. The final product will be either a ketone or an aldehyde.

For example, an aldehyde has one carbon-carbon bond and will react to form formic acid. Secondary alcohols will break 2 carbon-carbon bonds and will be oxidized twice, also forming formic acid. Primary alcohols will break one carbon-carbon bond and will be oxidized once to formaldehyde. Ketones will break two carbon-carbon bonds and form carbon dioxide (CO2).
Evidence that can be elucidated from periodic acid cleavage
[edit | edit source]This method can provide several clues to elucidate the structure of an unknown carbohydrate. By analyzing the ratios of the products mentioned above, some information about the types of functional groups present can be obtained. Specifically, because ketones oxidize to carbon dioxide when they are reacted with periodic acid, this method can provide clues as to whether the carbohydrate in question is an aldose or a ketose. Also, the size of the carbohydrate can be revealed by the amount of periodic acid that is consumed. One equivalent of periodic acid corresponds to one carbon-carbon bond cleavage. For example, a six-carbon carbohydrate would consume 5 equivalents of periodic acid.
Examples of periodic acid cleavage
[edit | edit source]One example of periodic acid cleavage involves the degradation of one equivalent of D-glucose into five equivalents of formic acid and one equivalent of formaldehyde.
Another example of periodic acid cleavage involves the degradation of one equivalent of D-fructose to three equivalents of formic acid, two equivalents of formaldehyde, and one equivalent of carbon dioxide. A ketose is a ketone with two or more hydroxyl groups (-OH), where at least one of the hydroxyl group at each end. Ketoses are a type of monosaccharide, which are important fuel molecules and nucleic acid building blocks. The simplest example of a ketose is dihydroxyacetone. It is a three-carbon structure containing one keto group and two hydroxyl groups (shown below). If you look closely at the image below you will notice that the keto group is in a slightly different color. The formula for this structure is CO(CH2OH)2. Ketoses also play important roles in the Calvin cycle, transaldolase reaction and transketolase reaction. Each of these processes are described in the following sections below.

The simplest ketose is Dihydroxyacetone. The stereochemical relations between D-ketoses containing three-four, five, and six carbon atoms are shown below in the family tree of ketoses:

Furanose Formation
[edit | edit source]Hemiketal or Hemiacetal
[edit | edit source]A Hemiketal is formed when a ketone react with an alcohol. The ‘-OR’ in alcohol attacks the oxygen in ketone, thus breaking the ‘C-O’ double bond. And the ‘H’ in alcohol bonds to the O.

Furanose Ring
[edit | edit source]Fructose chain cyclize when the -OH on C5 attacks ketone on C2 to form intramolecular hemiketal. It can form both 5 membered furanose ring or 6 membered pyranose ring. the Furanose ring makes the envelope ring form, with either C2 or C3 out of the plane; these are called C-2-endo and C-3-endo.
Ketoses in Reactions
[edit | edit source]Transketolase Reaction
[edit | edit source]The Transketolase reaction is very similar to the Transaldolase reaction. However, the Transketolase is different because it transfers a two carbon unit instead of Transaldolase's three carbon unit. Thiamine pyrophospate (TPP) ionizes so that it has a carbanion which is a negatively charged carbon. The importance of carbanion is that they can attack carbonyls, so that carbons are added in a sense to the nucleophile. TPP attacks a ketose substrate where it than releases the aldose product to yield an activated glycoaldehyde unit. An activated glycoaldehyde unit is an electron sink because of a positively charged nitrogen atom where a carbonyl of an aldose product is attacked and then separated after some electron movement. The importance of the transketolase reaction is that it is the mechanism that the enzyme TPP uses to change a ketose substrate to a ketose product that has a different group attached to it.
Transaldolase Reaction
[edit | edit source]The transaldolase reaction involves the transfer or a three carbon dihydroxyacetone unit from a ketose donor to an aldose acceptor. Unlike the transketolase reaction, transaldolase does not contain a prosthetic group; instead the reactions begins with a Schiff base formed between the carbonyl group of the ketose substrate and the amino group of a lysine residue at the active site of the enzyme. Next the Schiff base is protonated and the bond between C-3 and C-4 break which releases the aldose product. The leftover negative charge on the Schiff-base carbanion is stabilized by resonance while the positive charge on the nitrogen atom of the protonated Schiff base acts as the electron sink. The Schiff-base remains stable until a suitable aldose becomes bound which allows the dihydroxyacetone to react with the carbonyl group of the aldose and the ketose product is released from the lysine side chain via hydrolysis of the Schiff-base.
Transaldolase is a target of autoimmunity in patients with multiple sclerosis which is the selective destruction of oligodendrocytes that selectively expresses transaldolase in the brain.
Ketose in the Calvin Cycle
[edit | edit source]The Calvin cycle, or dark reactions, is one of the light-independent reactions. In the third phase of the this reaction, a five-carbon sugar is constructed from six-carbon and three-carbon sugars. A transketolase and an aldolase are the major factors in the rearrangement. The transketolase, which is in the pentose phosphate pathway, requires a coenzyme, thiamine pyrophosphate (TPP), to transfer a two-carbon unit from a ketose to an aldose. Whereas the transaldolase transfers a three-carbon unit from a ketose to an aldose.
In summary, transketolase first converts a six-carbon sugar and a three-carbon sugar into a four-carbon sugar and a five-carbon sugar. Then, aldolase combines the four-carbon product and a three-carbon sugar to form the seven-carbon sugar. This seven-carbon sugar then finally reacts with another three-carbon sugar to form two additional five-carbon sugars.
References
[edit | edit source]Berg, Jeremy, John Tyzmozcko, Lubert Stryer. Biochemistry
Berg, Jeremy, John Tyzmozcko, Lubert Stryer. Biochemistry Sixth Edition page 306
General information
[edit | edit source]An Aldose contains an aldehyde with two or more hydroxyl groups attached; one of the hydroxyl groups is at end opposite to the aldehyde. An Aldose is a type of monosaccharides, which is a chiral molecule that plays a key role in the development of nucleic acids. The two simplest forms of Aldoses are L- and D-Glyceraldehydes, which are three-carbon structures that each contain one aldehyde and two hydroxyl groups. The L and D symbols apply to the two different configurations of the asymmetric carbon farthest from the aldehyde group.

In the figure below, the common D-aldose sugars are shown:

Family Tree of D-Aldoses.jpg
Hemiacetal
[edit | edit source]The carbonyl group in aldehydes and ketones may react with one molecule of alcohol to form a hemiacetal. The ‘-OR’ group in alcohol attacks the carbonyl carbon in aldehydes or ketones, thus breaking the ‘C-O’ double bond. Proton transfer either intramolecularly or via solvent completes the reaction. Hemiacetal formation may be either acid or base catalyzed. Under acidic condition, however, the carbonyl group may react one more time when alcohol is in excess to form an acetal. The following two diagrams depict an example of hemiacetal and acetal found in carbohydrates.
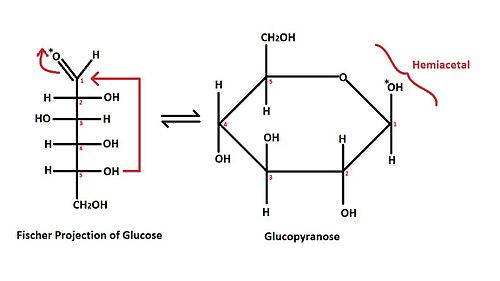

The red arrows shown in the Fischer projection of a glucose molecule demonstrate the brief overview of mechanism for the hemiacetal formation. The (*) mark on the oxygen of aldehyde indicates the position of the oxygen after the ring formation, while the numbers on each stereocenters indicate the position of each carbon.

Ring formation
[edit | edit source]Most of the sugars form cyclic rings, which are more stable than the open chain form. To enable ring formation, the aldehyde can react with an alcohol to form a hemiacetal. Carbohydrates may form either five or six membered rings depending on which hydroxyl group undergoes the hemiacetal formation. A five membered ring is called furanose, while a six membered ring is called pyranose. Furanoses form when the hydroxyl group on C4 reacts with the carbonyl group, while pyranoses form when the hydroxyl group on C5 reacts. This forms the intramolecular hemiacetal in the ring structure. In carbohydrates, the hemiacetal/acetal carbon (C1) in cyclic form is called the anomer. This carbon may be labeled as α or β depending on the position of the (*)-labeled oxygen in the figure. If the (*)-labeled oxygen in the picture is above the ring, the anomeric carbon is labeled as β. If below, it is labeled as α. The following is the formation of a five membered ring by a glucose molecule:

In other words, disaccharides are composed by 2 sugar molecules. They are called polysaccharides.
General information
[edit | edit source]Disaccharides, the simplest polysaccharides, are the products of a condensation reaction between two monosaccharides. Disaccharide is one of four groups of Carbohydrates (monosaccharide, disaccharide, polysaccharide, and oligosaccharide).

Formation
[edit | edit source]Disaccharides are formed when two monosaccharides join together by the dehydration synthesis reaction resulting in a glycosidic bond between the two monosaccharide molecules. The reaction produces water as a side product. The glycosidic bond in the picture below is a α-glycosidic bond because the bond is formed on the side opposite of the -CH2OH group.
Reducing Sugars
[edit | edit source]Most disaccharides are hemiacetals. Hemiacetals contain a free aldehyde to be oxidized into carboxylic acid. These are classified as reducing sugar. For example: maltose, lactose.
Carbohydrates that are acetals are not oxidized because both of its anomeric carbon atoms are fixed in a glycosidic bond. These are classified as non-reducing sugar. For example: Sucrose.

Classification
[edit | edit source]
There are two basic types of disaccharides: reducing disaccharides, in which disaccharides are Hemiacetals and contain a reactive carbonyl group, they are readily oxidized to diverse products.
Non-reducing disaccharides, in which the sugar is an acetals (or ketals) that cannot readily oxidized because both anomeric carbon atoms are fixed in a glycosidic linkage in which the components bond through their anomeric centers.
Properties
[edit | edit source]The glycosidic bond can form between hydroxyl groups on the two monosaccharides. Due to the different hydroxyl groups that bond, along with the alpha(α) or beta(β) position of the anomeric carbon, there are resulting disaccharides that are diastereomers differing in chemical and physical properties, depending on the monosaccharide components. The α-glucoside is more stable than β-glucoside due to anomeric effect. The C-R bond has a δ* antibonding orbital. If the C-R bond is in axial position, the antibonding δ* overlaps with one of the orbital of the oxygen, which stabilize the molecule. If the C-R bond is in equatorial position, there is no overlap between orbitals making the β-glucoside less stable than the α-glucoside.
Common disaccharides
[edit | edit source]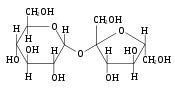


The most common disaccharides are Sucrose, Lactose, and Maltose.
Sucrose is the sugar often found in the grocery store and is produced by plants. It is a sugar derived from fructose and glucose. It is obtained from cane as a transport form of carbohydrates.
Lactose, found in milk, is formed by connecting β-D-galactose and α-D-glucose with a β-1,4-glycosidic bond.
Maltose is created by condensation reaction of the two glucoses, forming a α-1,4-O-glycosidic linkage. It is the second member of an important biochemical series of glucose chains. Maltose can be broken down into two glucose molecules by hydrolysis. In living organisms, the enzyme maltase can achieve this very rapidly.
Lactulose
[edit | edit source]galactose + fructose
Lactulose is a synthetic (man-made) sugar that is not absorbed by the body but is broken down in the colon into products that absorb water into the colon, thus softening stools.
Its primary use is to treat constipation. It is also used to reduce blood ammonia levels in persons with liver disease since lactulose absorbs ammonia into the colon (removing it from the body).
Trehalose
[edit | edit source]glucose + glucose
Trehalose is also known as tremalose or mycose. It is a natural alpha-linked disaccharide with extremely high water retention properties.
In nature, it helps plants and animals reduce long periods without water.
Cellobiose
[edit | edit source]glucose + glucose
Cellobiose is a hydrolysis product of cellulose or cellulose-rich materials, such as paper or cotton. It is formed by linking two beta-glucose molecules by a β(1→4) bond.
Table of Common Disaccharides
[edit | edit source]Here's a quick summary of the subunits of common disaccharides and how they are linked to each other.
| Dissacharide | First Unit | Second Unit | Bond |
|---|---|---|---|
| sucrose | glucose | fructose | α(1→2)β |
| lactulose | galactose | fructose | β(1→4) |
| lactose | galactose | glucose | β(1→4) |
| maltose | glucose | glucose | α(1→4) |
| trehalose | glucose | glucose | α(1→1)α |
| cellobiose | glucose | glucose | β(1→4) |
| chitobiose | glucosamine | glucosamine | β(1→4) |
There are many other disaccharides, although they are not as common, including isomaltose (2 glucose monomers), turanose (a glucose and a fructose monomer), melibiose (a galactose and a glucose monomer), xylobiose (two xylopyranose monomers), sophorose (2 glucose monomers), and mannobiose (2 mannose monomers).

Oligosaccharide is a carbohydrate polymers comprise three to ten monosaccharides, or, simple sugars. They were linked together mostly by O-glycosidic bond through condensation reaction between an anomeric carbon of a monosaccharide and the other. They can also form N-glycosidic linkages under certain atmosphere. The minimum numbers of reducing sugar components is one molecule lesser than the total number of simple sugars. Reducing sugar can be characterized from the hydroxyl group (-OH group) on the anomeric carbon.
Use of Oligosaccharides
[edit | edit source]Numbers of oligosaccharide molecules may form polysaccharides through multiple linkages between the anomeric carbon at the end of a molecule and the hydroxyl groups on another oligosaccharide molecule. Through O-glycosidic linkage and N-glycosidic bond, oligosaccharide may react with lipids and form lipopolysaccharides or saccharolipids. N-linked oligosaccharides can also react with the side chain(s) of amino acid residues - particularly Asparagine from a protein - to form a Glycoprotein. Glycoprotein does not form on random part of proteins. Glycoprotein usually forms on a residue that has sequence of Asn-X-Ser or Asn-X-Thr. However, not all of such residues will be attached to sugar molecule.
They are usually linked due to nitrogen or oxygen bonds to compatible amino acids. Oligosaccharides are known to be found in glycolipids and glycoproteins. Some of them are found from the breakdown of starch and cellulose, they are called cellodextrin and maltodextrin. Chemical marking is one of the functions of oligosaccharides, this is because they have much variations let similarities. For instance,blood types are marked by oligosaccharides. 'A' blood type has one oligosaccharide, 'B' blood type has one too. Both of these oligosaccharide markers are present in 'AB' blood type while 'O' blood type has none.The reason why blood needs to be typed before transfusion is because these oligosaccharides in blood are different enough than each other to be attacked by the body's immune system. Oligosaccharides can be identified as antigens detected by immune systems with incompatible blood as foreign pathogens. If transfusion were to occur with incompatible blood types, clotting and major illnesses would occur ultimately causing death. However, type AB blood contains all the possible oligosaccharide combinations possible (A or B) and since type O blood has no markers attached to it, type AB blood carriers are generally called universal acceptors. Whereas type O blood carriers can only accept blood transfusion from other type O blood donors which do not contain any oligosaccharides present within red blood cells.
Galactooligosaccharides
[edit | edit source]Galactooligosaccharides are synthesized through an enzymatic conversion of lactose. It is comprised of chain units of galactose group through consecutive transgalactosylation reactions, which the degree of polymerization ranging from 2 to 8 monomeric units. It is known that these new classes of prebiotics have an important role in improving gut health by sustaining beneficial and balanced gut microbiota. Today, many infant formula companies have galactooligosaccharides in their formula milk.
Human Milk Oligosaccharides
[edit | edit source]Human milk oligosaccharides are complex glycans that can be found in breast milk. One of the most important factors in infant’s diet is from breast milk, which pertains one of the most complex group of oligosaccharides known as Human milk oligosaccharides(HMOs). They are found in three, four, five, or even six chain sugars. For example, some of the HMOs include raffinose, 2’-fucosyl-lactose, 3’-fucosyl-lactose, 3’-sialyl-lactose, 6’-sialyl-lactose, and Lacto-N-tetraose. These HMOs differ in their size, structure, and specific linkages. There are more than 150 distinct Human Milk Oligosaccharide structures out there that are identified so far. Also, these HMOs are distinct in their structure, acidity, and functions. The backbone of Human Milk Oligosaccharides is the disaccharide lactose, which is formed by the linkage between galactose and glucose sugars. The final structure of HMO all depends on whether the backbone, lactose, is fucosylated or sialated, in either beta or alpha configurations or at a different carbon. For example, 2’-fucosyl-lactose has a fucose group at the alpha-1-3 position of the glucose monosaccharide of the lactose. Being sialated means the addition of a sialic acid group and formation of an acidic HMO.
HMO Formation
[edit | edit source]When the protein alpha-lactalbumin, also referred to as LALBA, is present, the enzyme beta-1,4-galactosyltransferase changes its function so it connects galactose to glucose sugars, which forms lactose. Human milk oligosaccharides are formed from lactose sugars, but the exact mechanism for this transformation is still unknown. HMOs are produced only when a woman is lactating and are formed in the mammary glands.
HMO Research
[edit | edit source]Colostrum, which is the liquid secreted by women’s breasts plus or minus several days of childbirth, are known to contain the highest amounts of HMOs, which varies per individual, but falls in the range of 20-30 g per liter. Colostrum contains more amounts of acidic HMOs relative to others. Matured breast milk contains a significantly less amount of approximately 10 g per liter. Therefore, premature newborns are fed breast milk that contains a higher amount of HMOs than would a baby born after 37 weeks of gestation, since the milk has not had the time to mature.
Although many medical professionals believe breast milk is a healthier alternative to feed newborns from HIV positive mothers than formula, recent findings (what sources ?) by call forth a problem. Studies have suggested that the presence of a specific sugar, 3’-sialyllactose, in breast milk has been found to increase the risk of a HIV-negative baby becoming infected with HIV from his or her HIV-positive mother. However, out of approximately 150 various HMOs, 3’-sialyllactose has been the only one to have any negative impact on the baby, while five others were determined to have a positive effect. Newborns who were fed breast milk that contained these five sugars lived longer than those who drank breast milk that did not contain the sugars. With more research on the effects of HMOs on babies, the findings could be applied to formula milk research. Since formula milk contains only small concentrations of complex oligosaccharides, as opposed to HMOs, studies can be conducted to determine if beneficial HMOs can safely be added to formula milk, and have the same positive effect on newborns. Also, women who produce breast milk containing high levels of 3’-sialyllactose can choose to feed their newborn formula, rather than breast milk. Overall, breast milk is the more beneficial choice to feed a newborn, over formula. Studies strongly indicate that HMOs decrease the likelihood of pathogens attacking the respiratory, urogenital, and gastrointestinal tracts of newborns.
Extracting HMOs
[edit | edit source]HMOs can successfully be isolated from breast milk. First the milk must be pasteurized to kill off any bacteria. Then the milk undergoes centrifugation to separate the expel the lipids from the aqueous phase. Afterwards, proteins can be formed into pellets, and then removed. Finally, the sugars are left, and they can be separated by gel permeation chromatography, which separates the sugars according to their masses. This is an example of size-exclusion chromatography that uses an organic solvent to elute the sugars.
External links
[edit | edit source]- Bode, Lars. "Recent Advances on Structure, Metabolism, and Function of Human Milk Oligosaccharides1." Journal of Nutrition 136.8 (2006): 2127-130. Web. 26 Oct. 2012.
- Engfer, Meike B., Bernd Stahl, Berndt Finke, Guenther Sawatzki, and Hannelore Daniel. "Human Milk Oligosaccharides Are Resistant to Enzymatic Hydrolysis in the Upper Gastrointestinal Tract 1." American Journal of Clinical Nutrition 71.6 (2000): 1589-596. Web. 26 Oct. 2012.
- www.bodelab.com
"Sequencing" an Oligosaccharide
Sequencing an Oligosaccharideinvolves identifying its structure. The most common method in doing so uses enzymes that cleave oligosaccharides at certain linkage points. The first step is to remove it from the protein it is attached to. From this point, the oligosaccharide can be isolated and the mass can be found using mass spectrometric techniques such as MALDI-TOF (Matrix Assisted Laser Desorption/Ionization Time-of-Flight). Unfortunately, since many possible oligosaccharide structures exist within a given mass. A better analysis can be obtained by further decomposing the oligosaccharide into smaller pieces using specific enzymes. The decomposition and analysis of the pieces of the oligosaccharide can be repeated as many times as possible or needed to find the structure of the oligosaccharide.


Polysaccharides are complex carbohydrate polymers consisting of more than 2 monosaccharides linked together covalently by glycosidic linkages in a condensation reaction. Being comparatively large macromolecules, polysaccharides are most often insoluble in water. Polysaccharides are extremely important in organisms for the purposes of energy storage and structural integrity.
There are two types of polysaccharides: homo-polysaccharides and hetero-polysaccharides. A homo-polysaccharide is defined to have only one type of monosaccharide repeating in the chain; whereas, a hetero-polysaccharide is composed of two or more types of monosaccharides. In both types of polysaccharide, the monosaccharide can link in a linear fashion or they can branch out into complex formations. It should also be noted that for a polysaccharide to be considered acidic it must contain one or more of the following groups: phosphate, sulfuric, or carboxyl.
Use of Polysaccharides
[edit | edit source]Polysaccharides have several roles. Polysaccharides such as starch, glycogen, and dextrans are all stored in the liver and muscles to be converted to energy for later use. Amylose and Amylopectin are polysaccharides of starch. Amylose has a linear chain structure made up of hundreds of glucose molecules that is linked by a alpha 1,4 glycosidic linkage. Due to the nature of these alpha 1,4 bonds, the macromolecule often assumes a bent shape. The starch molecules form a hollow helix that is suitable for easy energy access and storage. This gives starch a less fibrous quality and a more granule-like shape which is better suited for storage. Unlike the linear structure of Amylose, the Amylopectin starches are branched containing an alpha 1,6 glycosidic linkage about every 30 glucose units. Like amylose it is a homopolymer composed of many glucose units. Glycogen is found in animals, and it is branched like amylopectin. It is formed by mostly alpha 1,4 glycosidic linkages but branching occurs more frequently than in amylopectin as alpha 1,6 glycosidic linkages occur about every ten units. Other polysaccharides have structural functions. For example, cellulose is a major component in the structure of plants. Cellulose is made of repeating beta 1,4-glycosidic bonds. These beta 1,4-glycosidic bonds, unlike the alpha 1,4 glycosidic bonds, force cellulose to form long and sturdy straight chains that can interact with one another through hydrogen bonds to form fibers.
Polysaccharide Branching
[edit | edit source]Unbranched polysaccharides contain only alpha 1,4 linkages. However, there exists branched polysaccharides which are branched by virtue of certain molecules being linked to a molecule via alpha 1,4 and another via alpha 1,6 glycosidic bonds. The rate at which these bonds appear may vary. The plant based amylopectin contains a branch every 30 units while the animal based glycogen contains a branch approximately every 10 units. In digesting these branched polysaccharides, α-amylase is the relevant catalyst. α-amylase, however, only digests α-1,4 glycosidic bonds, leaving disaccharide/polysaccharide fragments containing α-1,6 bonds. These smaller fragments are known as Dextrins.
Glycogen and Starch
[edit | edit source]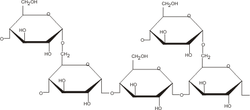
Many organisms store energy in the form of polysaccharides, commonly homopolymers of glucose. Glycogen, the polysaccharide used by animals to store energy, is composed of alpha-1,4-glycosidic bonds with branched alpha-1,6 bonds present at about every tenth monomer. Starch, used by plant cells, is similar in structure but exists in two forms: amylose is the helical form of starch comprised only of alpha-1,4 linkages, and amylopectin has a structure like glycogen except that the branched alpha-1,6 linkages are present on only about one in 30 monomers. These polysaccharides often contain tens of thousands of monomers, and each type is synthesized in the cell and broken down when energy is needed.
Glycogen metabolism is an intricate process involving many enzymes and cofactors resulting in the regular release and storage of glucose. This metabolic process is in turn broken down to glycogen degradation and synthesis. Glycogen synthesis is carried out by the enzyme glycogen synthase in which the activated form of glucose, UDP-glucose (uridine diphosphate), is formed by way of the reaction between UTP and glucose-1 phosphate. From this synthesis two outer phosphoryl groups are released from UTP producing the pyrophosphate compound. Pyrophosphate becomes an important aspect in this portion of the synthesis as the reaction to produce UDP-glucose is readily reversible. What allows the reaction to be driven forward is the hydrolysis of the pyrophosphate to orthophosphate in an irreversible reaction thus allowing the production of UDP-glucose to continue unhindered. The UDP-glucose is then attached to the non-reducing ends of glycogen. How this is accomplished is through an alpha-1,4-glycosidic linkage at the C-4 terminal with the terminal hydroxyl group ready to bind on glycogen. At this point the enzyme glycogen synthase plays the important role of catalyzing the attachment of UDP. Since an oligomer of at least four monomers is required for glycogen synthase to extend a chain, the process uses a primer that is itself provided by another enzyme, glycogenin. After several units of UDP have been attached to the glycogen by way of alpha-1,4 linkages, branching begins to take place by breaking an alpha-1,4 link and forming a alpha-1,6-link.A number of other enzymes, including insulin, play important roles in glycogen's synthesis. The breakdown of glycogen is completed through an entirely different biochemical pathway. Epinephrine and glucagon are signaling molecules whose binding to certain 7TM receptors activate the degradation, which is carried out in the cells by glycogen phosphorylase. This enzyme breaks up the polysaccharide chain by replacing the glycosidic bond with a phosphate group. As with its synthesis, glycogen's degradation requires numerous enzymes besides those mentioned here.
Starch is a good storage of carbohydrates because it is an intermediate compared to ATP and lipids in terms of energy. In plants, starch storage folds to allow more space inside cells. It is also insoluble in water, making it so that it can stay inside the plant without dissolving into the system. Starch can also be used as a back up source of energy when plants cannot obtain carbon dioxide, light, or nutrients from the surrounding soil.
Cellulose
[edit | edit source]Cellulose is the major polysaccharide found in plants responsible for structural role. It is one of the most naturally abundant organic compounds found on the planet. Cellulose is an unbranched polymer of glucose residues put together via beta-1,4 linkages, which allow the molecule to form long and straight chains. This straight chain conformation is ideal for the formation of strong fibers.
Although mammals cannot digest cellulose, it and other plant forms are necessary soluble fibers that mammals can eat. Pectin, for example, slows down the movement of food molecules in the digestive tract, which thereby allows for more necessary nutrients to be absorbed by the body instead of being quickly passed through as waste. Likewise, insoluble fibers like cellulose expedite the digestive movement of food molecules, which is imperative in the quick removal of harmful toxins.
Humans can't digest cellulose because we lack cellulases that would allow us to cleave the beta 1,4 linkages. However, some animals do eat and obtain energy from cellulose. One example of that is termites. These animals digest cellulose in a stepwise manner, using a combination of their own cellulases (produced in the foregut) and those of a microbial community resident in the distal parts of their digestive tract. This is a great example of a symbiotic relationship.
Cellulose is insoluble in water and aqueous solutions. It forms crystals and hydrogen bonds with amino acids. This quality of using intra and intermolecular hydrogen bonds to make crystals renders cellulose excessively insoluble in water and aqueous solutions. However, individual strands of cellulose aren't very hydrophobic as compared to other polysaccharides. It is the property of forming crystals that makes cellulose so insoluble.
Use of Cellulose
[edit | edit source]Cellulose has many uses, for example, as a gelling agent because of cellulose's properties of holding on to water. It is also used as an anticake agent, stabilizer, thickener and dispersing agent. Water cannot enter crystalline cellulose but dry cellulose absorbs water and it becomes flexible. Cellulose can give improved volume and texture particularly as a fat replacer in sauces and dressings but its insolubility means that all products will be cloudy.
Most papers are made of cellulose. The most important role of cellulose is that it is the major constituent of paper and cardboard and of textiles made from cotton, linen, and other plant fibers.
Cellulose can also be converted into cellophane, a thin transparent film, and into rayon, an important fiber that has been used for textiles since the beginning of the 20th century. Both cellophane and rayon are identical to cellulose in chemical structure. They are known as "regenerated cellulose fibers" and are usually made from viscose, a viscous solution made from celluloseare . A more recent and environmentally friendly method to produce rayon is the Lyocell process.
In the laboratory, cellulose is used as the stationary phase for TLC (thin layer chromatography). It is the raw material in the manufacture of nitrocellulose (cellulose nitrate) which was historically used in smokeless gunpowder and as the base material for celluloid used for photographic and movie films until the mid 1930s.
About a third of the world's production of purified cellulose is used as the base material for a number of water-soluble derivatives with pre-designed and wide-ranging properties dependent on groups involved and the degree of derivatization. Cellulose is also used to make hydrophilic and highly absorbent sponges. Derivatizing cellulose interferes with the orderly crystal-forming hydrogen bonding, described above, so that even hydrophobic derivatives may increase the apparent solubility in water. Methyl is thermogelling, which is made by methylating about 30% of the hydroxyl groups, forming gels above a critical temperature due to hydrophobic interactions between high-substituted regions and consequentially stabilized intermolecular hydrogen bonding. Such gels break down when cooling, in a manner similar to that causing the solubility minimum for non-polar gases; hydrophobic saccharides becoming less soluble as the temperature increases. This property is useful in forming films as barriers to water loss and for holding on to small gas bubbles.
Cellulose fibers are also used to make filter bed of inert material in liquid filtration.
Cellulase, an enzyme found in organisms that can digest cellulose, has recently garnered the attention of the scientific community and especially of alternative energy researchers who hope to produce biofuel (such as ethanol) by enzymatically breaking down the cellulose found in plants. How cellulase operates is not yet fully understood, but as more knowledge about the mechanism is uncovered researchers will be able to improve its efficiency (known cellulases are currently too slow to be used in industry) and apply its use to producing "green" energy sources. In this way, the most abundant source of bioenergy on Earth, cellulose, can become a part of the world's accessible energy supply. Some types of cellulase already find uses in industry, for example in food production and the textile industry.
Chitin
[edit | edit source]Chitin is a linear homopolysaccharide (long chain polymer) comprising N-acetyl-glucosamine, derivatives of glucose. These subunits form beta-glycosidic linkages similar to those formed by glucose molecules in cellulose. In fact, the only chemical difference from cellulose is the replacement of a hydroxyl group at C-2 with an acetylated amino group. Chitin can thus be described as cellulose, but simply with a different group at the second carbon. This increases hydrogen bonding, resulting in stronger molecules.
Chitin is the exoskeleton of many arthropods, and is the main component of cell walls in fungi, radulas of mollusks etc. Like cellulose, it is indigestible by vertebrate animals. Chitin has also been used as surgical thread, making it very valuable.
Glycosaminoglycan
[edit | edit source]Glycosaminoglycans (known as GAGs or mucopolysaccharides) are long unbranched polysaccharides made of repeating disaccharide units. Glycosaminoglycans are made from protein cores that are made in the endoplasmic reticulum that are posttranslationally modified by the Golgi. Here GAG disaccharides are added to protein cores to create proteoglycans.
GAGs are essential to life; they form important components of connective tissues and are found in collagen and elastin due to GAG chains that are covalently bonded to other proteins, forming proteoglycans. Because of this, water sticks to GAGs and, since water is not compressable, allows resistance to pressure. It is also used in smoke detectors due to its polarity.
Links
[edit | edit source]References
[edit | edit source]1. Berg, Jeremy M. 2007. Biochemistry. Sixth Ed. New York: W.H. Freeman. 310-323.
Introduction
[edit | edit source]Chitin is a nitrogen modified polysaccharide made up of N-acetylglucosamine, bound together in beta 1, 4 glucosidal bonds. Chitin is very similar in structure of cellulose; it has the additional amine component and hydroxyl substituent on each monomer. Chitin has an overall positive charge, making it great at binding to negatively charged objects, such as skin and proteins. It is insoluble in water and organic solvents. There are alpha, beta, and gamma forms of chitin. Alpha chitins are composed of alternating antiparallel polysaccharide strands mostly found in crustacean. Beta chitins are composed of parallel strands of polysaccharides, often found in squid. 2 parallel chains alternating with an antiparallel strand constitute gamma chitin and are found in fungi. One chitin derivative is chitosan, which is soluble in water and is made when chitin is placed in a chemical solution and well heated. Chitin has extensive medical uses, chitin when used during sutures increase healing by 50%, making the process shorter and less painful. Chitin can also be used for artificial blood vessels, antibacterial sponges, and dressings. Chitin can also act as a water purifier, it contains internal hooks which can remove impurities in water. In farming, chitin treated seeds tend to be resistant to fungus. In its unmodified form, chitin appears to be translucent, hard, and flexible. It mainly functions as the hard exterior of arthropods, and does a good job of protecting the soft interiors of these organisms from their harsh environment. It acts as structural support for organisms and prevents fluid loss in these organisms.
http://academic.brooklyn.cuny.edu/biology/bio4fv/page/chitin.jpg
[4]

Polysaccharides and oligosaccharides are also known as glycans. Glycans usually possess O-glycosidic linkages between monosaccharides. Cellulose, for example, is a glycan with β-1,4-linked D-glucose. Chitin is another glycan with β-1,4-linked N-acetyl-D-glucosamine. Glycans can be homo or heteropolymers of monosaccharide residues. They can have linear or branched features. Glycan may also refer to carbohydrate portions of glycoprotein, glycolipid, or a proteoglycan. Glycans can also be modified by a variety of different substituents, such as sulfation and acetylation. A variety of modifications of glycan enhances their diversity in nature and often serves as mediators of specific biological functions. For example, because many glycan are on the outer surface of cellular and secreted macromolecules, they are in charge of a wide variety of events in cell-cell, cell-matrix, and cell-molecule interactions that are important in the development and functions of a cell. Another function of these glycan is to act as a mediator in the interactions between different organisms.
Human Milk Oligosaccharides
[edit | edit source]Human milk oligosaccharides are complex glycans that can be found in breast milk. One of the most important factors in infant’s diet is from breast milk, which pertains one of the most complex group of oligosaccharides known as Human milk oligosaccharides(HMOs). They are found in three, four, five, or even six chain sugars. For example, some of the HMOs include raffinose, 2’-fucosyl-lactose, 3’-fucosyl-lactose, 3’-sialyl-lactose, 6’-sialyl-lactose, and Lacto-N-tetraose. These HMOs differ in their size, structure, and specific linkages. There are more than 150 distinct Human Milk Oligosaccharide structures out there that are identified so far. Also, these HMOs are distinct in their structure, acidity, and functions. The backbone of Human Milk Oligosaccharides is the disaccharide lactose, which is formed by the linkage between galactose and glucose sugars. The final structure of HMO all depends on whether the backbone, lactose, is fucosylated or sialated, in either beta or alpha configurations or at a different carbon. For example, 2’-fucosyl-lactose has a fucose group at the alpha-1-3 position of the glucose monosaccharide of the lactose. Being sialated means the addition of a sialic acid group and formation of an acidic HMO.
Introduction to Glycobiology
[edit | edit source]Chemical glycobiology deals with how glycans are formed and broken down. It deals with what glycan's biological roles are when they are settled and how the roles can be modified. To understand these issues, scientists have used a cooperative strategy of interrogation and perturbation. The interrogation strategy main purpose is to study and understand endogenous reactions and interactions between natural glycans and their binding complements. To be able to possess naturally forming and special glycans will enable researches to study protein-glycan and enzyme-glycan interactions. Arrangements made up of glycoconjugates, also known as lectins are useful tools for finding out more about protein-binding specificity or cellular glycosylation patterns. Along with the perturbation approach, using inhibitors, analaogs and substrates that are not natural can find out more about biosynthesis and how glycans function biologically. Both different oligosaccharides that are not natural and synthetically produced glycoconjugates can discourages or promote certain biomolecular interactions within the cells and organisms. In addition, there are compounds that are found to close off important steps to the process of the glycan biosynthetic pathways.
Another important subjects to cover is carbohydrate analogs, which are carbohydrates of similar structures with just different substituted groups. Carbohydrate analogs are used with glycans in many different ways. One example is imaging glycans. Another one is cross linking them to binding partners. Using these tools and chemical strategies, the molecular mechanisms that deal with glycan function can be better understood.
Glycan Synthesis
[edit | edit source]Pinpointing and defining oligosaccharides and glycoconjugates are important for understanding how glycans operate and function. To get these things from natural sources is very hard because when these substances produce, it deals with the interaction of multiple transporters and enzymes. This makes the whole process very complex. It is very complex as shown by the pathway for eukaryotic glycoprotein synthesis. The materials that form saccharides need to be produced and then sent to the correct cellular place. This place is where they are utilized for glycosyltransfereases. The speediness and optimization of making certain glycans are dependent upon how concentrated the building are, the type of glycosyltransferases and different biosynthetic enzymes in addition to the Km value of the these building blocks that are responsible for the glycosyltransferases that utilize them. The pathways that create N-glyocporteins, O-glycoproteins, glycolipids, glycosylphosphatidlyinositol anchors, proteoglycans, and polysacchardies are affected how easily the nucleotide donors can be obtained. However, the mechanism that control the governance of these pathways are currently in the process of being figured out. Therefore, is extremely hard to get enough amounts of glycans to examine and study from their biological sources.
Chemical strategies are now being utilize to deal with this issue. The strategy for dealing with this is providing the means to produce growning amounts of a variety of glycans. Glycans that occur naturally can be synthesized just like derivatives. This is significant because important relationships between the structure and activity can be examined further. Two basic strategies for synthesizing oligosaccharides are chemical and enzymatic. [1]
References
[edit | edit source]Introduction
[edit | edit source]A peptidoglycan biosynthetic pathway is a pathway involved in the production of the peptidoglycan layer, which makes up the cell wall in bacterial cells. The peptidoglycan layer is made up of glycan chains, which are cross-linked peptide substituents. As a main pathway for the production of this layer, the disruption in this pathway causes a weakened cell wall, or may even lead to bacterial lysis. Recent research has improved our understanding of peptidoglycan biosynthesis, which may potentially lead to the development of peptidoglycan-based therapeutics.[1]
UDP-N-Acetylmuramyl pentapeptide synthesis
[edit | edit source]The first step in peptidoglycan synthesis occurs in the cytoplasm with the murA-catalyzed transfer of enolpyruvyl moeitry from phosphoenolpyruvate(PEP) to uridine diphosphate-N-acetylglucosamine(UDP-GlcNAc). This transfer process is catalyzed by murA, and gram-negative bacteria only have one copy of this gene; therefore, its deletion is often lethal. Gram-positive bacteria, on the other hand, have two murA genes. The crystal structure of murA, in aoenzyme form and in complex with ligands, has been determined from several species of bacteria including E.coli. The structures contain two globular domains made up of four beta sheets and two parallel helices, and between these two domains is an active site, which is rearranged and brought closer together when substrates are attached. Fosfomycin, a naturally occurring antibiotic, inactivates murA by mimicking PEP and modifying the active site. [1]
The next step involves MurB catalyzing the NADPH-dependent conversion of enolpyruvyl UDP-GlcNAc to UDP-MurNAc in the cytoplasm. Following this production, a series of ATP-dependent amino acid ligases catalyze the addition of penetapeptide side chain onto this UDP-MurNAc complex. There are four additional Mur Ligases (Mur C, D, E, and F), and the structures of these enzymes have been determined. They all contain three structural domains that are involved in the peptide bond formation dependent on ATP hydrolysis. These domains include an N-terminal domain for binding the substrate, a centra ATPase domain, and a C-terminal domain to bind the amino acid.
MurC catalyzes the first addition of L-Alanine onto UDP-MurNAc, to which MurD adds the D-Glutamine. In the addition of L-Alanine, the C-terminal of the UDP-MurNAc substrate is activated by phosphorylation that requires ATP, and the intermediate that results from this is attacked by the animo group of the incoming L-Alanine. The intermediate is an acyl phosphate, and this acyl phosphate mechanism is also used by MurE in the addition of mesodiaminopimelic acid. MurF then adds D-Ala-D-Ala to the UDP-MurNAc-L-Ala-D-Glu-mesoA2pm, which is the product of MurE.
Assembly of Lipid II on the Inner Cytoplasmic Membrane
[edit | edit source]In the first membrane-associated step, an integral membrane protein named MraY transfers the MurNAc pentapeptide from the cytoplasm to a phosphate carrier. This results in a product known as lipid I. MraY is a type of a prenyl sugar transferase. Through the use of a thin-layer chromatography, the production of the lipid I product was able to be analyzed. It has been found that MurG, the final cytoplasmic component of the pathway, interacts with MraY to convert lipid I to lipid II via glycosyl transfer. High-resolution X-ray was used to show the structure of MurG and its domains.
References
[edit | edit source]Carbohydrates can be attached to proteins to form glycoproteins. In glycoproteins, the carbohydrate chains are either attached to the nitrogen atom in the side chain of asparagine (N-linkage) or to the oxygen in the side chain or serine or threonine (O-linkage.) There is also a third category of glycoproteins, non enzymatic glycosylated glycoproteins.
Glycoprotein Linkages[1]
[edit | edit source]N-Linked glycoproteins
[edit | edit source]In N-linked glycoproteins, an asparagine is only available for glycosylation if the residue is part of an Asn-X-Ser or Asn-X-Thr sequence (where X can be any residue.) While not all possible glycosylation sites are glycosylated, this specificity allows potential glycosylation sites to be detected.
Furthermore, all N-linked glycoproteins begin as an oligosaccharide attached to a specialized lipid molecule, dolichol phosphate, which resides in the ER membrane. As the oligosaccharide is transferred to the selected asparagine receptor, the structure is "flipped" through the ER membrane where additional sugars are then added by enzymes in the ER lumen. These N-linked glycoproteins are then transported to the Golgi complex where the carbohydrates can be modified to their final configurations.

O-Linked glycoproteins
[edit | edit source]Unlike the N-linked glycoproteins which arrive in the Golgi as a glycoprotein, O-linked glycoproteins have their sugar components developed solely within the Golgi complex. In the Golgi, the addition of a carbohydrate to the serine and threonine residue of a protein results in an O-linked glycoprotein.
Non-enzymatic glycosylated glycoproteins: synthesized by chemically adding sugar to the polypeptides.
Glycoprotein Function[2][3]
[edit | edit source]Structure
[edit | edit source]Glycoproteins can be found throughout matrices and act as receptors on cell surfaces that can then bring other cells and proteins (for example, collagen) together giving strength and support for a strong matrix system. For example, Proteoglycan-linking glycoproteins can cross link other proteoglycan molecules and create a formation of a ordered structure within the cartilage tissue. In nerve tissue, glycoproteins are also very abundant in the gray matter portion of the brain and appear to be associated with synaptosomes, axons, and other microsomes. Glycoproteins also play a huge role in the blood clotting mechanism because of their varying identities and roles as prothrombin, thrombin and fibrinogen. In addition, select bacteria contain a mucous like layer that surrounds the outermost components of their constituent cell walls which are also made of glycoproteins composed of very high molecular weight. These glycoproteins are commonly known as cellulose, also found in the structure of plants. The composition of such glycoproteins is important to the plant cell because the rigidity and the homogeneous form of such glycoproteins is what allows plants to stand upright. The flagella of bacteria are also made of structural glycoproteins in the sense that they are in bundles protruding from the cells surface, used to rotate and propel the cell in a specific direction.
Protection
[edit | edit source]Mucins are high molecular weight glycoproteins which can often be found protecting internal epithelial surfaces such as the respiratory, digestive, and urinary tracts of humans. Additionally, sweat glands also often secrete glycoproteins which can protect the skin.
Reproduction
[edit | edit source]Glycoproteins are important in the binding of a sperm cell to the surface of the egg during reproduction.
Hormones
[edit | edit source]HCG (Human chorionic gonadotropin) and EPO (erythropoietin) are both glycosylated proteins that function as hormones in the human body.
Due to the variety of glycoproteins, one being N linked while the O linked (pertaining to the amine terminus of Asn or the carboxyl terminus in Ser or Thr) both have different roles on oligosaccharide chains on the structure-function of glycoprotein hormones. In order for the differentiation of roles to be discovered on a hormonal level, an experiment was conducted in a fashion that O-glycosylation on the structure function of glycoprotein hormones were isolated and expressed in a CHO mutant cell line, 1dID, which specifically had a reversible defect in the protein o-glycosylation with only N-linked oligosaccharides being functional. The same was done for the N-glycosylation on the structure function of glycoprotein hormones excepted it was isolated into a different CHO mutant cell line which specifically had a reversible defect in the protein n-glycosylation with only o-linked oligosaccharides being functional. The results then indicated that the O-linked oligosaccharides played a very minor role in the receptor binding and signal transduction of the glycoprotein hormones dictated by the fact that the activity of the hormones remained unchanged. However the o-linked oligosaccharides did express a critical necessity for in vivo half-life and bioactivity. In other words, every other function that o-linked oligosaccharides previously had besides the assumed association with hormones was affected due to the absence of the glycoprotein. When N-linked oligosaccharide absence was analyzed, it was found that there was a development of glycoprotein hormone antagonists. In the specific case of hTS, the development of such antagonists can possibly offer therapeutic strategies in the treatment of thyrotoxicosis caused by Graves’ disease and TSH secreting pituitary adenoma.
Immunological Response
[edit | edit source]The specificity of different antibodies is determined by the carbohydrate structure in the glycoprotein. Additionally, both B and T cells contain surface glycoproteins which can bind certain antigens.
- ↑ Berg, Tymoczko, and Stryer (2002). Biochemistry. W.H. Freeeman and Company: New York. 5th edition: pg. 306-309.
- ↑ Ivatt, Raymond J. (1984) The Biology of Glycoproteins. Plenum Press: New York.
- ↑ Gottschalk, Alfred 1972 Glycoproteins: Their Composition, Structure, and Function. Elsevier Publishing Company: New York.
Fuad Fares, The role of O-linked and N-linked oligosaccharides on the structure–function of glycoprotein hormones: Development of agonists and antagonists, Biochimica et Biophysica Acta (BBA) - General Subjects, Volume 1760, Issue 4, April 2006, Pages 560–567, ISSN 0304-4165, 10.1016/j.bbagen.2005.12.022. Single units of carbohydrates called monosaccharides may undergo various reactions to form carbohydrate derivatives. Derivatives in case of carbohydrates often means modification of sugar molecules by addition substituents other than hydroxyl group. Amino sugars, acidic sugars, deoxy sugars, sugar alcohols, glycosylamines, and sugar phosphates are examples of carbohydrate derivatives. After modification, a transformed molecule of sugar resembles structure of a sugar and an added substituent; however, it is not considered as sugar anymore because its function and characteristics has changed. For instance, sugar alcohols still have sweet taste, but are not completely absorbed by the human body and, therefore, the impact of sugar alcohols on blood sugar is less and they provide fewer calories per gram. Thus, some sugar alcohols are widely used as sugar replacement in diet and health-oriented foods especially for individuals with diabetes. Most sugar derivatives occur naturally and have important biological functions. For instance, amino sugar heparin occurs in intracellular granules of mast cells that line arterial walls and, when released, inhibits blood clotting. Glycosylamine adenosine is an important part of DNA and RNA structure. Also, due to wide application of sugar derivatives in different areas many of them are commercially synthesized.
Glucose Family
[edit | edit source]



The Glucose Family are monosaccharides with functional groups replacing the hydroxyl group at the C-2 carbon or the C-6 group. Each sugar modification has a prominent effect in metabolism.
β-D-Glucose- This carbohydrate derivative plays a crucial role in metabolism. It is the main source of energy which is starts the process of cellular respiration. It is produced through photosynthesis and is often used in food products. It’s the most common carbohydrate and also circulates through the blood. It is known as blood sugar and can be converted into starch by the body. It is also used in blood tests for diabetes.
β-D-Glucose 6 Phosphate- Plays an important role in glycolysis. It is the final product after glucose is broken down and converted into the energy. The addition of the phosphate group gives the sugar a negative charge which prevents sugars from easily crossing lipid membranes. At elevated levels, it can inhibit brain hexokinase. It can be converted into starch or glycogen where it is stored in the liver and muscles. It plays an important role in blood glucose levels. Low levels can lead to Glucose-6-phosphate dehydrogenase deficiency, but the disease is hereditary. The disease leads to breakdown of red blood cells when exposed to certain environments.
β-D-Glucosamine- This is also an amino sugar that is very important in the formation of lipids and proteins. This can also form chitin in exoskeletons of insects as well as cell walls of plants. One of its common uses is for osteoarthritis. It helps rebuild cartilage and is used in veterinary medicine. Another of its uses is in helping with joint function and connective tissue. It also helps in body regulation and functions with β-D-Glucose 6 Phosphate in the body.
N-Acetyl- β-D-Glucosamine- This sugar is essential for optimal health and function in the body. It aids in cell communication. It also plays a role in how the immune system reacts with HIV and tumors. This also plays a role in osteoarthritis and helps in cartilage formation. It has been shown to play a role in nerve functioning for learning in mammals. This molecule has multiple uses such as limiting cholesterol absorption and decreases insulin secretion. Some receptors have been found in the thyroid to transport iodine proteins. It is found in multiple glands of the body and plays some role in the organ’s function. Muramic Acid-This sugar is a main component of bacteria cell walls and it is a derivative of peptidoglycan. It is used in gas chromatography for laboratory experiments.
N-Acetylmuramic Acid- This is also found in peptidoglycans of bacterial cell walls. It is a the product after N-acetylglucosamine that has been condensed with lactic acid. In bacteria, phosphophoenolpyruvate adds the lactyl group to C3 of N-acetylglucosamine. Beta-lactams compete for binding with transpeptidases, enzymes that catalyze the formation bonds between N-acetylmuramic acid-based peptide chains. Chlamydia lacks N-acetylmuramic acid in its cell walls which is the reason why penicillin does not treat the disease.
β-D-Glucuronate-This molecule is a highly polar molecule. It is used to increase solubility of some drugs. It is incorporated into proteoglycans and then combined with steroid hormones. This molecule forms to help make compounds more soluble for excretion. Bilirubin is one of the main molecules that this molecule makes soluble. This compound is very useful in clearing drugs from the body and making them soluble so that drugs can be processed through the body.
β-D-Gluconate – This is used as an alkalinizing agent for fluid therapy. It can be used in cleaning products because it dissolves mineral deposits. An alkaline solution will aid in dissolving the minerals faster. It can slowly be metabolized into bicarbonate, but the effects are longer lasting. It is an acidic sugar that occurs naturally in plants, honey, and fruits. It can be prepared through fermentation of glucose as well. When combined with calcium, it can form a gel to treat burns that are caused by hydrofluoric acid.
Amino Sugars
[edit | edit source]


Amino sugars are generally known as monosaccharide carbohydrate sugars that have replaced an -NH2 amine group with the 2'-carbon hydroxyl substituent. The most abundant amino sugar is one of the oldest and most abundant organic compounds on Earth. More than 60 amino sugars are known, many of them having been isolated and identified only recently as components of antibiotics. Examples of amino sugars include:
Galactosamine, for example, is one of eight essential amino acids that function in cell to cell interaction. Though research on galactosamine has just begun, research has shown that it may help those with joint inflammations. Lacking in galactosamine may even be one of the factors related to heart disease. Also, it may also function as a toxin leading to liver failure. In addition, galactosamine may also function as one of the sugars composing the follicle stimulating hormone (FSH) and the luteinizing hormone (LH), both of which are needed in the reproductive processes of the human body. Sources of galactosamine include bovine (which includes both cattle and oxen), red algae and shark meat.
Glucosamine, contrary to galactosamine is a very popular and relatively well-known amino sugar that our bodies use to produce glycoconjugates like glycosylated lipids and proteins. Glucosamine has a structural role in composing the hard exoskeleton of chitins, such as a variety of arachnids, crustaceans, and insects. Our bodies can obtain glucosamine from external sources such as the fermentation of different grains, like wheat and rice and barley, and like galactosamine, from bovine and shark. As far as its effect on bodily functions, glucosamine's precursor, glucosaminoglycans are major segments involved in joint cartilage, and thus can help in the treatment of osteoporosis, or osteoarthritis.
Sialic Acid is a very important sugar amine necessary to our bodies' mental and physical well being. Without sialic acid, our bodies would deteriorate in both aspects. For example, children found lacking in this particular sugar amine have been known to be adversely affected in their development, growth and in the pigmentation of their hair and skin. In a study done by Bing Wang et al., improved sialic acid concentrations in infants proved to improve their synaptogenesis and their neurological development. However, although sialic acid may serve to help us, it may also be the culprit that allows different viruses to enter. Sialic acid is normally found on the surfaces of cell membranes, it is not only responsible for fluid uptake to its negatively charged regions, but it also has the ability to recognize and bind to proteins on certain viruses, such as Hemagglutinin which is found on the cell surface of the Influenza virus. Thus, once the two bind together, it opens a door for the virus to enter into the cell and spread to other parts of the body, causing injurious repercussions.
'N-Acetyl-d-glucosamine' is the main component of the polysaccharide in chitin, the substance that makes up the tough outer skeleton of arthropods and insects.
Deoxy Sugars
[edit | edit source]


Deoxy sugars are carbohydrate derivatives that are lacking an -OH hydroxyl group at the 2'-Carbon of the sugar cyclic ring, hence "deoxy," de- being a prefix meaning "to remove," and "oxy" representing "oxygen."
Deoxyribose is the most commonly known deoxy sugar because it is the exact sugar used in the backbone of our DNA double helices. Scientists speculate that the deoxyribose sugar derivative was used as opposed to ribose because of its more stable structure. Unlike ribose, it doesn't contain a hydroxyl group at its 2'-Carbon, which would otherwise make it susceptible to hydrogen bonding with other species or molecules, thus disrupting the kind of stable matrix that our DNA needs to have. In the DNA backbone, deoxyribose sugars are bound to phosphate groups via phosphodiester linkages, and are each covalently attached to one of the four DNA nitrogenous bases. They therefore play very important roles in in the flexibility of the DNA backbone and in ensuring that the polar nitrogenous bases are facing into the double helix rather than outwards where they would be unstably exposed to hydrophobic media.
Fucose, although less talked about in our science books, is also an essential sugar derivative needed to maintain our bodies' overall well-being and proper development. Fucose is known to be excreted by nursing mothers, and in recent studies has shown to have a significant role in the fetal development of newborns, as well as to the advancement of their immune systems. Fucose and its derivatives also ensure the proper transmission of nerve signals or cell to cell communication, improve the brain's long-term memory and even play an important role in inhibiting the spread of tumor and cancerous cells.
Rhamnose is not as well known, and for a while scientists had trouble studying this sugar and understanding what its purpose was in the body. In fact, it used to be thought of as an inert deoxy sugar. It is also unusual in the sense that unlike most other sugars, its natural occurring form is its L configuration (i.e. L-Rhamnose as opposed to D-Rhamnose). Interestingly enough, Rhamnose sugar can be extracted from poison sumac. Generally, rhamnose is found mostly in the cells of plants and bacteria rather than animals. Rhamnose may participate in the body in cell proliferation, collagen synthesis and in the degradation of free radicals.
Dideoxy and Trideoxy Sugars
Dideoxy sugars and trideoxy sugars are sugars where two or three of the hydroxyl groups, respectively, are replaced with hydrogen atoms. These sugars are most commonly made by plants, fungi and bacteria. Over one hundred different sugars of these types have been isolated from prokaryotic sources alone.
Di and trideoxy sugars are found attached to the lipopolysaccharides that are embedded in the outer membrane of Gram-negative bacteria, as well as in other areas of these bacteria’s S-layer. They are also found on the S-layer of Gram-positive bacteria, which is the outer peptidoglycan membrane which is thicker in Gram-positive bacteria than in Gram-negative bacteria. In medicine, these rare sugars are used in antibiotic, antifungal, anthelmintic and antitumor agents, and are therefore quite valuable to science.
'Their Synthesis'
The enzymes concerned with their production are largely unknown, mainly because these unusual dideoxy and trideoxy sugars are not commercially available, so not much study has gone on concerning them thus far. However, the overall method of their synthesis has been observed. In the biosynthesis of these dideoxysugars and trideoxysugars, one of the most important intermediates to form is NDP-4-keto-6-deoxyglucose (where NDP stands for nucleotidyl diphosphate). This molecule serves as the branching point for hundreds of different di and trideoxy sugars. Its synthesis undergoes two basic steps (illustrated in Steps 1 and 2).
- In Step 1, α-D-glucose-1-phosphate is attached to an NMP moiety using a nucleotidyltransferase type enzyme. Which particular enzyme depends on which nucleotide is included in the reaction, as each nucleotide has a unique shape.
- In Step 2, the carbon number 6 is reduced, removing the hydroxyl to give a CH3 instead of the original CH2OH. Also, the carbon number 4 hydroxyl group is oxidized, leaving one double-bonded oxygen in its place. This step is catalyzed by another enzyme given the name NDP-glucose-4,6-dehydratase.
- Step 3 shows seven enzymatic reaction types commonly utilized to form the plethora of possible di and trideoxy sugars (see illustration). These are the reactions where many of the enzymes involved still remain unknown. Their discovery and determinations of their structures and more detailed function is the next step for biochemists in the field of dideoxy and trideoxysugars.
Acidic Sugars
[edit | edit source]

Acidic Sugars- According to Brønsted and Lowry, an acid is defined as any molecule that has the ability to donate one of its hydrogens (protons). It is this losing of a proton that makes a molecule acidic. As far as sugars go, a sugar's simplest structure is called a monosaccharide (if you think of it in SAT terms: atom is to element as monosaccharide is to sugar), and what makes a sugar acidic is the oxidation of one of its -OH groups after a hydrogen (proton) has been "lost" turning the carbon into a carbonyl carbon (carboxyl group) making an acid out of the molecule.
Some Of The Most Commonly Found Acidic Sugars
[edit | edit source]Glucuronic Acid: Glucuronic acid derives from Alpha D-glucose by means of oxidation. Mechanism can be found here [6](image drawn by author using a chemistry drawing program from https://scifinder.cas.org). The most common function of glucuronic acid is in the metabolism of foreign compounds (i.e. medication or poisons) in an organism. This molecule is extremely important to the chemists who make and design drugs as it is pertinent to know if and how a medication will be metabolized in the body. Glucuronic acid in Greek is called "sweet urine" which is a sugar that exists in urine. The acid functions to bind together toxins, such as drugs, hormones and steroids, found in the liver, helping them flush out from the body. Glucuronic acid is often used as a detoxicating agent which can help with drug overdosing and minimizing drug interaction within the body. Recent studies also show how Glucuronic Acid prevents prostate cancer in men by detoxing chemicals within the body. Ascorbic Acid: Ascorbic acid is most commonly known by the name Vitamin C (the L-enantiomer of ascorbic acid). Because of its ease in being able to oxidize, vitamin C has been used for such things as a preservative. It is also known as a cure for scurvy (a lack of vitamin C in the body). This was most common back in the day when sailors or pirates would go out on long trips on the ocean, their supply of fruit would expire and they would have no source of vitamin C and thus get scurvy. Since human bodies cannot produce vitamin C on their own, we must be able to get our vitamin C from the food and fruits that we eat. Ascorbic Acid also has antioxidant properties which can help protect nucleic acids, proteins, lipids, and other cell organelles from free radicals such as hydroxyl radicals (reactive oxygen molecules) that could else wise be damaging and tumor-inducing. The oxidized version of ascorbic acid is relatively safe, unreactive, and can be metabolized without any problems. However, an excess amount of ascorbate, which is an oxidized ascorbic acid molecule, could potentially promote and initiate instead of limiting free radical reactions within a living system.
Artificial Sweeteners
[edit | edit source]Artificial sweeteners provide the same, if not more, sweetness of the sugar without the high calories or subsequent tooth decay that accompanies sugar consumption.
Aspartame
[edit | edit source]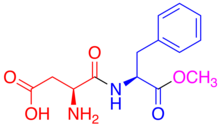
This sugar substitute, known on the market as NutraSweet (for baking purposes) or Equal, was discovered in 1965, but not approved by the FDA until 1981. It is 180 times as sweet as sucrose and contains 4 calories per gram, which is considered negligible. It is unstable in heat and decomposes in liquid during prolonged storage. However, although the aspartame may decompose in liquid, it is not hazardous for consumption. The break-down of aspartame only affects the quality of the beverage. Aspartame is a dipeptide consisting of two amino acids in an ester bond with methanol. Aspartic acid is at the N-terminus and phenylalanine is at the C-terminus where it bonds with methanol. Aspartame is metabolized in the gastrointestinal tract, where the peptide and ester bonds are broken, separating the amino acids and the methanol. The effects of the increase in methanol concentration from the breakdown of aspartame in the body was studied by Soffritti, et al. in Italy. This group studied rats and the effect of high dosages of aspartame in linkage to lymphomas and leukemia. It was found that lymphomas and leukemia increased in the female rats at dosages around the acceptable daily intake (ADI). And the levels increased in male rats only at the highest dosage which was 100 times the ADI. They also noticed that increasing the methanol intake through water increased the leukemia, as did the addition of methyl-ter-butyl ether (MTBE) which metabolizes to produce methanol. Methanol in the body turns into formaldehyde which can turn into formic acid. The study also showed that increasing the amount of formaldehyde increased the leukemia and lymphomas. While this study seems to indicate that aspartame consumption is a risky health hazard, there is much criticism for the study. For one, the group did not allow another group to examine their samples which goes against the customary procedure of verification. In addition, the FDA has criticized this lab in the past for conducting unreliable work.
While the validity of aspartame being a health hazard is being questioned, it is known for a fact that aspartame is quite hazardous to individuals with Phenylketonuria (PKU). Those with PKU cannot metabolize phenylalanine which causes a toxic amount of phenylalanine to accumulate in the body if substances that contain phenylalanine, like aspartame, are consumed.
Saccharin
[edit | edit source]
Also known as Sweet-N-Low, saccharin is one of the oldest artificial sweeteners. It is 300 times as sweet as sucrose. Saccharin was discovered at Johns Hopkins University in 1879 in the course of research on coal-tar derivatives. Its name, which comes from the Latin word for sugar, saccharin, has no structural relationship to any sugar. Saccharin itself isn't very soluble in water. However, the proton bonded to nitrogen is fairly acidic and saccharin is normally marketed as its water-soluble sodium or calcium salt. Its earliest applications were in as a replacement for sugar in the diet of diabetics before insulin became widely available. The enhanced sweetness permits less to be used and therefore reducing the cost of production. Using less carbohydrate-based sweetener also reduces the number of calories.
Sucralose
[edit | edit source]
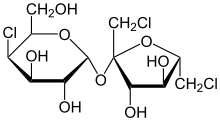
More popularly known as Splenda, sucralose is 600 times sweeter than sucrose. It is very similar to the disaccharide structure of sucrose. Three of the hydroxyl groups are simply replaced by chloride atoms. Although this change does make the molecule more likely to react with nucleophiles and could present as a danger due to the addition of chlorine, it is not a cause for concern. The levels of chlorine are no different than in comparison to consumption of table salt which is made of sodium chloride, and is harmless in moderation. Sucralose is considered calorie-free because the body does not to recognize the molecule as sugar, so it does not get broken down for energy.
References & External links
[edit | edit source]Carey, Francis A., and Neil T. Allison. "23." Organic Chemistry. 8th ed. New York: McGraw-Hill, 2011. 1049. Print. Carey, Francis A., and Robert M. Giuliano. "23.12 Amino Sugars." Organic Chemistry. 8th ed. New York: McGraw-Hill, 2011. N. pag. Print.
http://www.drugs.com/dict/d-glucose-6-phosphate.html
http://www.revolutionhealth.com/healthy-living/vitamin-index/n-acetyl-d-glucosamine-ns
http://www.glyconutrient.biz/N-AcetylGlucosamine.htm
http://themedicalbiochemistrypage.org/non-glucose-sugar-metabolism.html
Meister, K. “Sugar Substitutes and Your Health.” Comprehensive Reviews in Food Science and Food Safety, 2006.
Soffritti, M, et. al. "Aspartame Induces Lymphomas and Leukaemias in Rats" Eur. J. Oncol., vol.10, 2005.
Introduction
Many proteins either have carbohydrates added on to them for structural and purposes or use carbohydrates as substrates in cell reactions.
CARBOHYDRATES BINDING MOLECULES AND MALARIA
In a paper titled Carbohydrate Binding Molecules in Malaria Pathology Alan Brown and Matthew K Higgins studied the relationship between glycoproteins and the malaria parasite.
-
Plasmodium falciparum ring-forms and gametocytes in human blood.
-
Main symptoms of malaria.
-
The life cycle of malaria parasites in the human body.
-
Anopheles albimanus mosquito feeding on a human arm.
Many pathogens, such as bacteria, viruses, and parasites, use carbohydrate-binding molecules to interact with their hosts.Carbohydrates play an important role in the modification and function of proteins.The new insights on pathogenic carbohydrate-binding molecules provide potential therapeutic targets.Glycoproteins play a key role in pathogen binding to their host cells. Malaria-causing parasites, interact with human hepatocytes and erythrocytes. This glycoprotein mediated binding is essential for host cell invasion, movement, avoidance of detection by the immune system, and destruction.
The malaria parasite recognizes multiple sialylated proteins on the erythrocyte surface to bind to the host cell. Two protein families in malaria parasites recognize the sialic acid on the erythrocytes: EBL (erythrocyte-binding like) and RBL (reticulocyte-binding like). Both protein families consists of proteins that interact with different membrane protein and lead to different pathways; their function can be either independent or dependent of sialic acid, which enables the pathogen to overcome the diversities among human population.Both these proteins contain a region called RII. This region is responsible for the ability of the malaria protein to bind to the host cells. EBL and RBL have multiple methods of binding and attack on erythrocytes. This means that it can bind in multiple different ways and avoid destruction by the human immune system.
EBL Family
EBL family is composed of RII region (made of two Duffy-binding like (DBL) domains known as F1 and F2), a C-terminal cysteinerich region and a transmembrane domain.RII region is conservative; it reveals 98% sequence identity across strains from various geographical locations.One of the family members, EBA-175 is better understood by the researchers.
EBA-175
EBA-175 binds to sialylated membrane glycoprotein of erythrocyte, Glycophorin A. Its importance in invasion pathway has been shown through in vitro experiment where antibodies against EBA-175 inhibit binding to Glycophorin A and block invasion.The RII region EBA-175 recognizes and binds to Glycoprotein A, a glycoprotein on the surface of the host cell. Glycoprotein A has specific carbohydrate sequences on its surface that are essential for binding to occur. When the malaria parasite binds to Glycoprotein A, it recognizes sialiac acid on the cell’s surface. This is a good method of attack for the parasite as it is unlikely that sialic acid compositions changes. However, slight differences in sialic acid can prohibit attack by the parasite.
Nevertheless, sialic acid alone is insufficient for pathogenic invasion. For instance, Glycophorin B, which shares 11 conserved sialylated O-linked glycans with Glycophorin A, does not bind EBA-175. In fact, certain orientation of the protein chain is required for optimal binding, where EBA-175 is more likely to directly interact with the protein.
In its crystal form, the two DBL domains of RII region are joined through a three helical linker where two anti-parallel monomers form a dimer. Such structure consists two positively charged channels. Each channels lined with residues from the F1 domain of one monomer and the F2 domain of the other. In solution, though, EBA-175 appears to be concentration dependent in which content ratio of monomer and dimer varies with solute concentration. Thus, it can be proposed that EBA-175 is stable as a monomer and tends to dimerize at high concentrations.
By co-crystallize EBA-175 with Neu5Ac(a2,3)Gal, researchers are able to detect sugar molecule, locating near the channels of dimer, are coordinated by residues of both monomers. This observation aid in the efforts to locate sialic acid binding site and results in the model where EBA-175 assembles around the Glycophorin A ectodomain (glycoproteins on the surface of the membrane and extends into extracellular space) by interactions with both sialic acid and the protein chain.
There are also alternative invasion pathways that involve other members from EBL family. In particular, EBA-140 is devoid of charged patches and residues identified as interacting with sialic acid in EBA-175, while it still binds to Glycophorin C in a sialic acid dependent way. Therefore, each member of EBL family seems to have its own sialic acid binding site distinct from others.
Cytoadhesion and Evasion of Host Detection
Infected erythrocytes become sticky and adhere to uninfected erythrocytes to form clumps known as rosettes. This prevents detection and spleen mediated destruction of infected cells allowing the infection to continue to survive in the body. In Malaria targeted pregnant women, the erythrocytes will accumulate and lead to placental inflammation, possibly causing loss of the child. Antibodies that bind to PfEMP1proteins and prevent adhesion in some forms of Malaria suggests PfEMP1proteins as potential therapeutic targets.PfEMP1. These PfEMP1 proteins are made of DBL (duffy binding-like) domains.
PfEMP1proteins The PfEMP1 proteins contain large-sized ectodomains made from multiple DBL and CIDR domains. Different PfEMP1 proteins will interact with different receptors. Among PfEMP1 proteins, VAR2CSA is the best understood by the researchers.PfEMP1s switch rapidly to avoid detection in the body. These interact with various proteins (CD36 and ICAM-1) as well as carbohydrates. One of these PfEMPS1 is VAR2CSA.
VAR2CSA
The infected erythrocytes by VAR2SCA become specifically avid for 4-sulphated chondroitin sulphate A (CSA) molecules. CSA molecules, especially those that are closely related glycosaminoglycan carbohydrates, show little ability to prevent interactions with VAR2CSA. In addition, the protein chain of CSA molecule does not contribute to the specificity of recognition of the molecule.Structurally, Four out of the six DBL domains from VAR2CSA bind to CSA in isolation. The DBL3X structure has an a-helical structure similar to the EBA-175 DBL domains, only with longer loops.
When sulphate ions or disaccharides are present, a loop on the concave surface of the domain will form a positively charged patch and sulphate-binding pocket crucial for CSA binding. Different form EBA-175, VAR2CSA has longer, often disordered, loops and is more versatile. The versatility of VAR2CSA hints that VAR2CSA has a longer length of exposure to immune system in comparison to EBA-175 during invasion process. The structure of the DBL6e domain is similar toDBL3X, but it does not have the sulphate-binding pocket or positively charged patch. It binds to CSA molecule through positive charges on a distinct surface of the domain, a mechanism that shows more versatility than DBL3X. Therefore, it is more likely to be a more exposed part of the intact ectodomain during cell invasion.
The two domains have distinct surface features to interact with CAS molecules. Moreover, they do not have the CSA-binding specificity; rather their affinity for carbohydrates increases with the charge of the carbohydrate. In addition, the ectodomain shows strong ligand specificity where the ectodomain acts as a compacted monomer distinct fromEBA-175.
Conclusion
Carbohydrate binding molecules are crucial in the life cycle of Malaria pathogen. Both EBA-175 and VAR2CSA use the same DBL domain in binding carbohydrates where the specificity of binding requires multiple domains. EBA-175 needs both protein and carbohydrate components of a dimeric receptor for specificity whereas VAR2CSA requires only carbohydrate for specificity; EBA-175 uses “carbohydrate binding surfaces lining the grooves at dimerization interfaces” whereas VAR2CSA uses “multiple domains from a single monomer to generate a specific binding pocket”. Studies of the carbohydrate-binding proteins and deeper understanding of their structures can provide useful information on pathogenic pathway as well as potential therapeutic targets in the future.
Introduction
[edit | edit source]
Lectins are proteins that recognize and bind specific carbohydrates found on the surfaces of cells. They play a role in interactions and communication between cells typically for recognition. Carbohydrates on the surface of one cell bind to the binding sites of lectins on the surface of another cell. Binding results from numerous weak interactions which come together to form a strong attraction. A lectin usually contains two or more binding sites for carbohydrate units. In addition, the carbohydrate-binding specificity of a certain lectin is determined by the amino acid residues that bind the carbohydrate. Lectins are specific carbohydrate-binding proteins: - Enormous diversity of carbohydrates have biological significance: Different monosaccharides can be joined to one another through any several -OH groups. Extensive branching is possible. Many more different oligosaccharides can be formed from 4sugars than oligopeptides from 4 amino acids - Lectins promote interactions between cells: Lectin is to facilitate cell-cell contact Lectin and carbohydrates are linked by a number of weak non-covalent interactions C-type(calcium required): calcium ion on the protein acts a bridge between protein and sugar through direct interactions with sugar -OH groups Carbohydrates-binding specificity of a particular lectin is determined by the amino acid residues that bind the carbohydrates. - Influenza virus binds to Sialic acid residues: Influenza virus recognizes sialic acid residues linked to galactose residues that are present on cell-surface glycoproteins. These carbohydrates are bound to hemagglutinin, a viral protein (virus is engulfed by the cell and starts to replicate). Neuraminidases are enzymes that cleave the glycosidic bond to sialic acid residues of hemagglutin, releasing the virus to infect new cells and spreading the infection.
Lectin Binding
[edit | edit source]Lectins are capable of binding to many different types of carbohydrates. Because of this capability, the way that a lectin binds to carbohydrates, the materials necessary for binding, and the strength of the bond varies. Some of the various forms of binding are discussed below.
-Monosaccharides and disaccharides have shallow grooves to which lectins bind, making the affinity of the bond low. Because of the difficulty that lectins face when binding to these carbohydrates, a subsite multivalency (which is a spatial extension of the grooves) is necessary to achieve binding. This extension makes it so that the contact site on the carbohydrate is embedded into a more complex contact region. This type of binding works most efficiently with small lectins, as evidenced by the lectin, hevein, which is only 43 amino acids long. Rapid binding kinetics also facilitate the binding of lectins to carbohydrates. An example of this is the binding of sialyl Lewisx (a tetrasaccharide) to P-selectin. Rapid binding kinetics allows for spatial complementarity to be reached between a low-energy conformation of the carbohydrate and the prearranged binding site of the lectin.
-The shape of the binding sites in carbohydrates plays a factor in its bondage to lectins. An example of this is the case of galectin-1 binding to ganglioside GM1 (a pentasaccharide). Nuclear Magnetic Resonance and other molecular modeling techniques were used to analyze the bond between these two molecules. The images found showed that two branches of the carbohydrate are bonded to the lectin. The α2, 3-sialylgalactose linkage is able to adopt three different, low-energy conformers. One of these conformers is energetically favorable for the binding of galectin- to ganglioside GM1. This process is evidence that lectins prefer certain conformations (shapes) when deciding how to bind to a carbohydrate. This evidence shows that oligosaccharides have limited flexibility. This limited flexibility makes oligosaccharides very favorable ligands, seeing as they avoid entropic penalties.
-Core substitutions have been found to occur in N-glycans. These substitutions are added to specific positions on the carbohydrate during its course to being assembled. These substitutions have been found to prominently affect the properties of glycans. The glycan properties are so affected, that they do not even need to be in the presence of lectins in order to be noticed. These substitutions, resulting in changes of certain parts of the carbohydrate, act as molecular switches governing the shape of glycans.
-Branching also introduces molecular switches. This property is most exemplified in the glycoside cluster effect. Enhancing the numerical valency of a molecule results in an increase in affinity. The type of branching appears to have a significant effect on this increase in affinity.
Importance of Carbohydrates in Cell Communication
[edit | edit source]Carbohydrates contain abundant information as a result of the various composition and structures that are possible. These diverse compounds result from the many OH groups available for linkage, which further allow for extensive branching. Additionally, the substituent attached to the anomeric carbon can assume either an alpha or beta configuration. The presence of these various carbohydrates on cell surfaces allows for effective cell-to-cell communication.
Functions of Lectins
[edit | edit source]Lectins are known to be very widespread in nature. They can bind to soluble carbohydrates or carbohydrate functional groups that are a part of a glycoprotein or glycolipid. Lectins typically bind these carbohydrates with certain animal cells and sometimes results in glycoconjugate precipitation.
In animals, lectins regulate the cell adhesion to glycoprotein synthesis, control protein levels in blood, and bind soluble extracellular and intracellular glycoproteins. Also, in the immune system, lectins recognize carbohydrates found specifically on pathogens, or those that are not recognizable on host cells. Clinically, purified lectins can be used to identify glycolipids and glycoproteins on an individual's red blood cells for blood typing.
C-Type Lectins
[edit | edit source]C-Type lectins are those that require a calcium ion. The calcium ion helps bind the protein and carbohydrate by interacting with the OH groups found on the carbohydrate. Calcium can also form a linkage between the carbohydrate and glutamates in the lectin. Binding is further strengthened through hydrogen bonds that form between the lectin side chains and the OH groups of the carbohydrate. Carbohydrate recognition and binding is made possible by a homologous domain consisting of 120 amino acids. These amino acids determine the specificity of carbohydrate binding.
C Type lectins carry a wide range of functions such as cell to cell adhesion, immune response to foreign bodies and self-cell destruction. C Type lectins are categorized into various different subgroups specific to the different protein functional domains. These lectins are calcium ion dependent and share linear structural homology in their carbohydrate-recognition domains. Among Eukaryotes and the animal kingdom, this wide range of protein families including endocytic receptors, collectins, and selectins is found most abundantly. The differences in members of the family vary in the different kinds of carbohydrate complexes that are recognized with high polarity and affinity. C type lectins are involved with immune defense mechanisms and help protect an organism against tumorous cells.
P-Type Lectins
[edit | edit source][1]
P-Type lectins contain a phosphate group. CD-MPR and CI-MPR are the only two members of the P-lectin family, cation-dependent and cation-independent. The main function of P-type lectins in eukaryotic cells involves delivering newly synthesized soluble acid hydrolyses to the lysosome. They do this by binding to mannose 6-phosphate residues found on the N-linked oligosaccharides of the hydrolyses.
MPRs (Mannose-6-phosphate receptors) were discovered when studies on mucolipidosis II, a lysosomal storage disorder, were conducted. Hickman and Neufeld found that fibroblasts from ML II patients were able to absorb lysosomal enzymes excreted by normal cells, whereas fibroblasts from normal patients were not able to absorb the lysosomal enzymes. Hickman and Neufeld hypothesized that the lysosomal enzymes had a recognition tag that allowed for receptor-mediated uptake and transport to lysosomes. These tags later became known as MPRs.
CI-MPR is about 300 kDA and exists as a dimer. The overall folding of CI-MPR is similar to that of CD-MPR, but unlike CD-MPR, CI-MPR is cation-independent. In addition, CI-MPR binds to proteins that have the MPR tag, IFG-II (a peptide hormone), and other non lysosomal hydrolases. The N-terminal three domains of CI-MPR exists as a monomer, and forms a tri-lobed disk that has significant contact with one another. This attribute of the tri-lobed disk is vital in maintaining the structure of its sugar binding site. Phosphorylated Glycan Microarray demonstrates that CI-MPR shows little disparity between glycans having one or two phosphomonoesters when it comes to binding. This is unlike CD-MPR, which has been shown to have affinity towards glycans with two phosphomonoesters. In addition, CI-MPR binds to ligands at the cell surface, unlike CD-MPR. Overall, all of the ligand binding sites of CI-MPR are located on the odd-numbered domains. Four signature residues in CD-MPR and domain 3 of CI-MPR are conserved, and have been found to react with Man-6-P in the same manner, suggesting that the Man-6-P binding pockets are similar. One difference that has been found is the fact that the pocket in CD-MPR contains Mn 2+, whereas the binding pocket in CI-MPR does not. This could be the reason why CI-MPR is cation-independent.
CD-MPR is a 46 kDA cation-dependent homodimer. Three disulfide linkages formed by six cysteine residues in the extracellular region of CD-MPR are key to the folding of the homodimer. Because the 15 contiguous domains of the extrasystolic region are similar in size and amino acid sequence when compared to each other, it is understood that CD-MPR and CI-MPR have similar tertiary structures. In fact, CD-MPR domains 1, 2, 3, 11, 12, 13 and 14 of CI-MPR have the same fold in the extrasystolic domain. The overall fold of the CD-MPR monomer consists of a flattened beta barrel consisting of two antiparallel beta sheets, one with four strands, and the other with five strands. The CD-MPR dimer consists of two five stranded antiparallel beta sheets. E133, Y143, Q66, and R111 have been found to be essential in Man-6-P binding via mutagenesis studies of CD-MPR. CD-MPR’s binding and unbinding mechanism is similar to that of the oxy-to-deoxy transition of hemoglobin. The overall movement has been described as to be a “scissoring and twisting” motion in between the two subunits of the dimer interface. These two subunits are connected via a salt bridge. Absence of this salt bridge results in a weaker bind with lysosomal enzymes, signaling the importance of ionic interactions between the two subunits in binding.
Selectins
[edit | edit source]Selectins are a type of C-Type lectins that play a role in the immune system. Selectins consist of L, E, and P forms that bind to carbohydrates found on lymph-node vessels, endothelium, and activated blood platelets. They behave analogously to C type lectins in that both have a high affinity for calcium binding and are responsible for immune responses. Selectins are sugar binding polymers that are adhesive among other cells which causes it to be highly effective in targeting an inflammatory response for a localized region. Selectins target only specific kinds of binding sites, but thus allows it to be effective in conjunction with leukocyte cascading to minimize invasively targeting an infected region.
Examples of Lectins
[edit | edit source]Embryos are attached to the endometrium of the uterus through L-Selectin. This activates a signal to allow for implantation.
E. coli are able to reside in the gastrointestinal tract by lectins that recognize carbohydrates in the intestines.
The influenza virus contains hemagglutinin which recognizes sialic acid residues on the glycoproteins located on the surface of the host cell. This allows the virus to attach and gain entry into the host cell.
References
[edit | edit source]Gabius, Hans-Joachim, Sabine Andre, Jesus Jimenez-Barbero, Antonio Romero, and Dolores Solis. "From Lectin Structure to Functional Glycomics: Principles of the Sugar Code." Trends in Biochemical Sciences 36.6 (2011): 298-313. Print.
Glycosaminoglycans (Muccopolysaccharides)
Structure
Glycosaminoglycans (formerly known as muccopolysaccharides) are disaccharides consisting of two hexose derivatives in a repeating, un-branched chain. The hexose derivatives are most commonly glucosamine or galactosamine. Along with being a hexose derivative, either a carboxylate or sulfate group is attached to one or both monomers of the disaccharide, giving it a negative charge. Some of the most important glycosaminoglycans include chondroitin sulfate, dermatan sulfate, keratan sulfate, heparin, heparan sulfate, and hyaluronate.
-
Some common glycosaminoglycans
Function
Glycosaminoglycans are essential molecules in the body. They can covalently connect to proteins in order to form proteoglycans. Proteoglycans act more as carbohydrates than proteins as most of their molecular weight is due to glycosaminoglycans (95%). These proteoglycans are integral parts of connective tissue such as tendons and cartilage, they can act as anticoagulants, and they are also a component of the fluid that lubricates joints.
Regulation
Glycosaminoglycans are regulated in the lysosome of the cell by being broken down in stages using specific enzymes. When those enzymes are not present in the lysosome, the undigested fragments build up in the cell, leading to what is referred to as a lysosomal storage disease. The class of lysosomal storage diseases involving glycosaminoglycans is called muccopolysaccharidosis (MPS). The types of MPS range from type 1-10 depending on the enzyme that is not present. This class of diseases can be debilitating and result mental retardation and loss of muscle function.
References
Biochemistry. 5th edition.Berg JM, Tymoczko JL, Stryer L.New York: W H Freeman; 2002
Proteoglycans are proteins that are covalently bonded at multiple sites along the protein chain to a class of polysaccharides, known as glycosaminoglycans. Glycosaminoglycans constitute approximately 95% of the mass of proteoglycans by weight, which results in proteoglycans bearing a resemblance more to polysaccharides than to proteins. The physiological properties of proteoglycans are a function of the particular glycosaminoglycans present. Examples of common glycosaminoglycans are chondroitin 6-sulfate, keratan sulfate, heparin, dermatan sulfate, and hyaluronate. As a result of the ionic character of glycosaminoglycans, proteoglycans carry at least one negatively charged carboxylate or sulfate functional group under physiological conditions[1]. Examples of proteoglycans include Versican, Brevican, Neurocan, and Aggrecan.

Synthesis
[edit | edit source]As the name implies, proteoglycans are essentially proteins that have been glycosylated. These proteins are synthesized in the Rough Endoplasmic Reticulum by ribosomes, and are then transported by vesicles to the Golgi Apparatus, where the proteins are modified into various forms. Proteoglycans are among these modified forms. Upon completion of synthesis, the proteoglycans are transported to the Extracellular Matrix by vesicles. [2]
Function
[edit | edit source]Proteoglycans are a significant component in the Extracellular Matrix. The major functions/purpose of proteoglycans depends on the glycosaminoglycan component of the molecule. This component allows connective tissues of the Extracellular Matrix (ECM) to be able to withstand compressional forces through hydration and swelling pressure to the tissue. Aggrecan best portrays this particular function.
In addition to Aggrecan are other proteoglycans such as Lumican, Decorin, Perlecan, Fibromodulin and Biglycan, each serving different functions according to their components. [3]
Examples
[edit | edit source]Aggrecan
[edit | edit source]In biological systems, proteoglycans constitute the structural building blocks of connective tissue; additionally, proteoglycans serve as joint lubricants. The most well studied proteoglycan, called aggrecan, is found alongside collagen as the main components of cartilage. The negative charges on the repeating glycosaminoglycan units of aggrecan attract water absorption. As a result, aggrecan acts as a cushion for impact retention by absorbing and desorbing water[4]. This role is particularly important in between joints that sustain high amounts of impact in mammalian bodies, such as the knees. Aggrecan has also been found to take an important role in the central nervous system. It is one of many proteoglycans that inhibit neuronal growth, movement, and interaction. These proteoglycans are important for keeping the pathways established in young, developing animal brains during adulthood; however they are also the reason adult brains are less likely to establish new connections than the developing brain. Recent work has hinted that aggrecan is an important contributor to aging neuronal loss and dementia[5].
Lumican
[edit | edit source]Lumican belongs to a family of proteoglycans called Small Leucine-Rich Proteoglycans. It is most well known for its importance in healing and scar tissue formation in mammals. Recent studies in mice show that it specifically helps mediate the migration of chemokines (cytokines responsible for preventing infection and promoting wound healing) to injured sites in the skin. Mice that lack the gene encoding named LUM for encoding the protein Lumican have tissue defects and poor immune responses to infections[6]. Lumican's role in healing injured sites is not completely understood, however the protein's moiety allows collagen fibrils and charged hydrophilic gylcosaminogyclans to bind into interfibrillar spacings. Lumican as a keratan sulfate proteoglycan forms interstitial collagenous matrices within an injured site, which not only aids in wound closing, but protection of the inflammatory site against infections. In addition, Lumican regulates collagent organization, spatial growth, corneal transparency, cell migration, and tissue repair.
References
[edit | edit source][1] Berg, Jeremy M., Tymoczko, John L., and Stryer, Lubert. Biochemistry. 6th ed. New York, N.Y.: W.H. Freeman and Company, 2007: 312, 313.
[2] Proteoglycan. (n.d.). In Wikipedia. Retrieved November 17, 2009, from http://en.wikipedia.org/wiki/Proteoglycan.
[3] Yanagishita, M. (1993). Function of proteoglycans in the extracellular matrix. Acta Pathol Jpn, 43(6), 283-93.
[4] Tanaka, Y. (2009). Influence of aging on chondroitin sulfate proteoglycan expression and neural stem/progenitor cells in rat brain and improving effects of a herbal medicine, yokukansan. Neuroscience, 164(3), 1224-.
[5] Lee, S, & Lee, S. (2009). Extracellular Matrix Lumican Deposited on the Surface of Neutrophils Promotes Migration by Binding to β2 Integrin. The Journal of biological chemistry, 284(35), 23662-.
Definition of Blood
[edit | edit source]- A type of complicated and living tissue which holds numerous proteins and cell types.
- Blood is important in our bodies due to its functions: defend, regulate, and transport. It is also known as fluid tissue present in blood vessels
Structure
[edit | edit source]Human blood groups depends on the functioning of glycosyltransferases, enzymes that catalyze the formation of glycosidic bond between the Structure and function of the human blood. Specific oligosaccharide antigens attach to the proteins and lipids on the surface of erythrocytes. Those attached to proteins have a serine or threonine residue or ceramide lipid intermediate. The most basic oligosaccharide attached is called the O antigen (also referred to as the H antigen). This O antigen is the base oligosaccharide found in all three blood types AB, A, and B. The O antigen is of the form (—Lipid—Glucose—Galactose—N-acetylglucosamine—Galactose—Fucose). Blood type O only has the O antigen attached to the red blood cells. Blood type A is formed through the addition of the A antigen, which has N-acetylgalactosamine (GalNAc) glycosidically bonded to the O antigen. Similarly for blood type B, the B antigen has an additional galactose forming a glycosidic bond to the O antigen. In both the A and B blood types, the new antigen forms an α-1,3 linkage to the outermost galactose component of the O antigen through the help of glycosyltransferases. GalNAc transferase adds the extra N-acetylgalactosamine for the A antigen while Gal transferase adds the extra galactose for the B antigen. Genes in a person’s DNA code for the specific glycosyltransferases to allow for the addition of antigens A and/or B to the O antigen. If a person’s genes do not allow for the coding of type A or B transferase, then that person will have type O blood. The genes that code for GalNAc and Gal transferases are exact, but for three amino acids. This strong similarity between the two enzymes shows they are related through divergent evolution.
Functionality
[edit | edit source]The importance of glycosyltransferases is most prominent when a person needs a blood transfusion. Type O blood can be given to anyone because everyone has the ability to recognize the O antigen. However, a person with type A or type O blood cannot recognize the B antigen while another with type B or O blood cannot recognize the A antigen. Antibodies against the B and A antigens are also present in the serum of one’s blood for those who lack the B and A antigens, respectively. Therefore, if a person were transfused with the wrong type of blood, one’s immune system will see the antigen as foreign, therefore attacking those transfused red blood cells. It is because of this process that people with type AB blood can accept transfusions of any blood type, since they have all three antigens already present in their bodies. It should be noted that in reality, transfusions are complicated by the Rhesus factor.
Proteins & Blood Types
[edit | edit source]- We can categorize our blood types into 4 groups: A, B, AB, & O
These four groups are considered to be the four phenotypes that one may posses. These four phenotypes can produce six different genotypes that each person can be: AA, AO, BB, BO, AB, and OO.
- On the surface of red blood cells, there are 2 types of antigen: A & B (each type has its own properties)
If a blood cell is type A, the surface of the cell contains Antigens for type A and the body will produce antibodies for type B and vice versa for type B. Type AB contains both antigens on the surface and has neither antibodies. Blood type O has no antigens and thus have both A and B antibodies in its system. In this regard, blood type O may be the universal donor and blood type AB is the universal recipient.
- We can also look at the property of Rh protein whether it's absence or presence in our blood.
- If a blood type has Rh protein, then it is positive.
- If a blood type does not have Rh protein, then it is negative.
- For example, blood type "B negative" means that the person has type B without Rh protein on the surface of the red blood cells.
The RH factor is more important if the mother is to have a second child. This is because if the mother is RH – and her first child is RH + (has present of RH protein), the mother will produce antibodies (specific immunogenic proteins) for the RH protein. This will not affect the first child since the mother will only produce this antibody after the child is born. However, if the mother is to have a second child with an RH+ trait as well, the antibodies in the mother will cross the placenta and attack the child. This is because the antibodies for the RH protein are specific proteins called memory cells that are small enough to pass the placenta. This will result in a miscarriage.
One way around this is to give the mother Rhogam after birth. This is a synthetic protein antibody that will destroy the left over fetal cells before it has a chance to interact with the mother's immune system. This inhibit protein antibody production towards the RH factor in the mother and will allow subsequent RH+ children to be born from that mother.
Mixing Different Blood Types
[edit | edit source]- Our body generates antigens or antibodies to protect us from the unfamiliar molecules. They will then recognize this difference and clash with the molecules to get rid of them.
- For blood transfusion, it is very important to make sure that both the recipient and the donor match in blood type.
- If surface molecules from the donor blood cells signals any difference than the recipient's, then the antibodies from the recipient’s blood will consider it as foreign.
- The immune response will take place if there is a difference in blood type, which results in blood clots in the vessels.
Universal Donors and Universal Recipients
- Blood type O is the universal donors due to its versatility of having no molecules on the red blood cell surfaces, which will not trigger any immune response. Therefore type O blood can be donated to any of the other four blood types earning its name.
- Blood type AB is the universal recipients due to its lack of antibodies that recognize type A or B surface molecules. AB can receive blood from any of the other four blood types earning its name as the universal receiver. AB however also tends to be the most rare blood type out of the four.
AB+ is the true universal receiver able to receive all types of blood regardless of type and Rh antigen.
However O- is only compatible with itself but able to donate to everyone.
The exception to these rules are people with the hh antigen system also called the Bombay blood type. These individuals cannot express the H antigen which is present in group O. They cannot make A antigen or B antigen since they are made from the H antigen. Therefore the people who have this blood type can donate to any other member of any blood type but can only receive blood from other Bombay blood type individuals. However this blood type is extremely rare occurring in only .0004% of the population. Therefore people with this blood type are at a great risk in finding compatible blood for a blood transfusion.
Rhesus Antigen
Not only do the different blood types have to be considered due to the existence of antigens but the rhesus blood group system is secondly important after the ABO system of blood type antigens. The most important antigen from the five main rhesus antigens is RhD since it is the most immunogenic. It is common for RhD negative people to have no anti-RhD IgG or IgM antibodies. The Rhesus antigen is usually depicted by a plus or a negative after the type of blood type. Rh positive is more prevalent than Rh negative blood types. Especially in East Asia the percentage of Rh negative people are extremely rare. Since people with Rh negative blood type cannot receive blood from Rh positive special care must be taken when receiving blood transfusions.
Testing for Rhesus Antigen
[edit | edit source]Test can be carried out to determine whether one's blood contains the Rh antigen. Since Rh factors are antigens there are corresponding antibodies that can be used to bind. Rh antibodies are commonly used to bind to such antigens in the blood. The structure of the antibody is oriented in a fashion so that there are two main chains, a heavy chain and light chain. The two heavy chains are located on the inner side of the antibody while the light chains are located on the outer. The domains are exactly the same pertaining to all antibodies except for the last domain the N terminus of both the heavy and light chain located on each side of the antibody. It is the variable domain that is different from antibody to antibody and contains differential amino acid sequences with their constituent residues that provide complementarity in the binding site to the specific substrate. If Rh antibodies are mixed with ones blood and aggregation occurs, then the person would be Rh positive, if no aggregation occurs, then the person is Rh negative. Aggregation occurs due to the antibody-substrate complex that forms when the Rh antigen locks into the binding site of the antibody.
Red Blood Cells Vs Blood Plasma
Like red blood cell compatibility recipients can receive blood plasma from the same blood type. However unlike with red blood cells the plasma has a converse compatibility. Blood type O can receive plasma from every other blood type while blood type AB can donate blood plasma to any blood type.
Blood Types Genetics
[edit | edit source]- Blood types A & B co-dominate
- Blood type O is recessive
- We can see the patterns of the possible combination of alleles versus the blood types:
AB = blood type AB
BB = blood type B
AA = blood type A
OO = blood type O
BO = blood type B
AO = blood type A
Blood type A can receive both type A and type O blood. Similarly, blood type B can receive both type B and type O blood. However, blood type AB can receive type A and type B blood, as well as, type O blood, making them the universal recipients. Blood type O is rare in that it can only receive type O blood, but can donate to any of the other three blood types, making them the universal donors.
References
[edit | edit source]Nelson, David L. Principles of Biochemistry, 4th ed. W. H. Freeman, 2004.
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?indexed=google&rid=mcb.section.4816 Some viruses enter their host cells by attaching to cell-surface carbohydrates. The influenza virus in particular, attaches to sialic acid residues on the terminai of the oligosaccharides present near the cell-surface glycoproteins and glycolipids.

Once attached, viruses inject their own genetic material and take over the cell's machinery to produce more viruses. The cell can undergo a lytic or lysogenic cycle. In the lytic cell, the cell is taken over, produces viruses with its own machinery and organelles, and then dies, releasing more viruses. In the lysogenic cycle, the cell does not die but instead replicates with viral DNA/RNA in its own genome. A common example of a lysogenic virus is lambda phage. However, lambda phage can also enter the lytic cycle. In this manner, some viruses can remain dormant for years at a time, activating under certain environmental conditions to begin replication under the lytic cycle.
There are several different types of viruses, including DNA, RNA, and retroviruses. DNA viruses usually replicate by taking over the cell's DNA, whereas RNA viruses tend to replicate in the cytoplasm. Retroviruses transcribe their DNA into host DNA via reverse transcription- an example of this is HIV.
The body has several defense mechanisms against viruses. The body's first line of defense is the innate immune response, which is non-specific and defends the body against a variety of threats. Inflammation, coughing, sneezing, and a variety of other non-specific defense mechanisms are examples of the innate immune response in action. The adaptive immune system, on the other hand, targets specific threats. T Cell and antibody activation are examples of the adaptive immune system's defense mechanisms. In addition, scientists have developed vaccines and anti-viral drugs to assist the immune system and/or disrupt viral replication mechanisms.
Lymphocytes, or white blood cells, are the body's main defenders against foreign invaders. There are two major types of lymphocytes, T cells and B cells. Of the T cells, there are T helper, cytotoxic, memory, regulatory, and natural killer cells. Each type of T cell has its own purpose in immune response. Helper cells assist and activate other cells at the first signs of infection, cytotoxic T cells destroy infected cells, preventing more viruses from being released, memory cells store antigens, allowing the body to quickly recognize previous infections to make fighting future infections more efficient, regulatory T cells shut down immune response after an infection has subsided, and natural killer cells can help kill tumor cells. B cells, on the other hand, recognize antigens and develop anti-bodies once the correct antigen for an infection has been found.
Viruses are parasites because they can't survive on their own. They infect a cell and use the infected cell to multiply and make more viruses. Viruses are considered alive because they are capable of duplication and have defensive mechanisms. All viruses have some sort of genome, which can be single or double stranded, DNA or RNA, and linear or circular. A virus genome is also very small: they could be four genes or a few hundred when prokaryotes tend to have several thousands of genes and eukaryotes tend to have tens of thousands of genes. Viruses have a protein coat that protects the genome, and this coat is made up of protein. Usually, the coat has multiple copies of one protein because the virus wants to minimize its genome. Viruses want to pack up and ship off new copies of viruses as soon as possible, so if the genome is longer and more varied, then the longer the packing will take because it will have to encode for several different genes; thus, viruses would be at a great disadvantage if they had variety in their genome.
What is A Virus?
[edit | edit source]A virus is an infectious, parasitic agent that can only replicate inside other host cells. All viruses have a genome and a capsid.
A viral genome can be:
•Single stranded (ss) or double stranded (ds)
•DNA or RNA
•Linear or circular
Compared to others, the viral genome is very small. It consists of two to several hundred genes. Prokaryotes, like bacteria, have thousands of genes, while eukaryotes can have tens of thousands of genes.
Viruses were once thought to have been a self-replicating genetic element but the guiding principle which differentiates the two is that “once a replicon incorporates a gene(s) that allows it to make a capsid to enclose the replicon, then a functional and structural entity called a virion is produced.” This means that the hallmark of a functional virus is a virion. [9]
The capsid is a protein coat that surround the genome. It consists of several copies of a single protein, which is advantageous for the virus because it only needs one gene to code for the capsid protein. The capsid and it's enclosed genome is also referred to as a nucleocapsid.
Some viruses also have a membrane bilayer around the nucleocapsid that can serve as a protective layer. This can be found in viruses that have already infected a host cell, and have budded off from the host cell.
Viral proteins have structures that are different from eukaryotic proteins in that they are loosely packed due to relatively fewer hydrogen bonding and van der Waals interactions. [2]
Viral proteins are small proteins with many disordered segments and many coil residues. The coil residues cause the protein to be loosely packed because the coil conformation (a secondary structure) is unorganized and does not easily lie near another protein strand. [2]
In addition, viruses are not considered living organisms as they fail to fulfill the basic requirements of life to be considered living. Although viruses replicate, they are not able to metabolize food. Viruses do not have an organized cell structure, it does not respond to any external stimuli when placed under such circumstances, nor do they maintain homeostasis in which internal temperature is attempted to be kept at a constant. Additionally, viruses do not belong to any animal kingdom, they are unable to adapt to environments, and their only method of reproduction is through invading host cells, but not being able to reproduce independently.
Origin of Viruses
[edit | edit source]The exact origin of viruses has been questioned by scientists for decades. Although there are many theories on the origin, there are three hypotheses that have been discussed at greater lengths about the origin of viruses.
1. Viruses originated from ancient times before cellular life was invented This hypothesis has been rejected many based on the idea that viruses need a cellular host in order to survive. If viruses existed before cellular life existed then they would not have had the ability to replicate in a host cell.
2. Viruses originated from cells by reduction. This hypothesis is rejected on the premise that this method requires intermediates between cells and viruses which have yet to be found.
3. Viruses escaped from cells by utilizing cellular replication elements removed from cellular control. This hypothesis doesn't explain how it is possible to build complex virion structures. Also since it has been found that viruses are capable to infecting cells in all three domains, it has been proven that they are ancient and not from reduced cells. Also the discovery of the mimivirus somewhat refutes this hypothesis.
When comparing a two different plant viruses, scientists were able to see the similarity in capsid structure and similarities in the arrangement within the capsid. When comparing two animal viruses, they were not only found to be similar to each other but also to plant viruses based on their coat protein folds. They all shared the stranded beta barrel folding. Plant, insect, and human viruses showed to share similar structural architecture which therefore led to the conclusion that ssRNA viruses may share a common ancestor. In addition to the ssRNA, after more comparisons, scientists saw that even dsDNA and bacterial viruses shared the same basic architect as ssRNA viruses; they all used the beta barrel fold. By comparing various viruses, it has been discovered that a common ancestor infected cells of all three domains before diversification. [9]
Virus Classification
[edit | edit source]Viruses are classified under two different classifications: the Baltimore Scheme and the International Committee on Taxonomy of Virus(ICTV).
The Baltimore Scheme divides all the various viruses into seven groups depending on their chemical type, number of strands, and if their single strand is capable of directly undergoing translation. The seven categories of viruses are dsDNA, ssDNA, dsRNA, plus(sense) ssRNA, minus(antisense) ssRNA, ssRNA with DNA intermediate, and dsDNA with RNA intermediate.
The second classification categorizes viruses into order, family, genus, species. The organization of the virus lineages is based on the information on the type of host cell each virus infects. Although the ICTV try to organize the viruses to the best of their ability, it is still difficult to designate a majority of the families to orders. [9]
Viral Structures
[edit | edit source]Viral structures have been found to be the unifying component within the viral universe. Viruses have several set structures and do not deviate from these structures. They can be icosahedral, helical, a combination of the two, bottle-shaped, lemon-shaped, and spindle shaped. Some viruses are also pleomorphic; they have a lipid envelope.
For classification purposes, the capsid structure has been declared to be the crucial element. The process of replication and the genomic structure were considered as an element in classification but because of the lack of information they provide for identity, it has been ruled out. The capsid structure is the main structural component that is congruent to the identity of the virus. [9]
Viral Reproduction
[edit | edit source]Viruses are obligate (imposed by necessity; incapable of adaptation to different conditions; restricted to a particular mode of life) intracellular parasites. They reproduce only within a host cell. Viruses contain no metabolic enzymes or "machinery" for protein synthesis. They can only infect a limited range/type of host cell (some viruses can infect several species, such as the rabies virus; some viruses can only infect one species; and animal viruses are usually tissue or cell-type specific; that is they will only infect one particular type of cell). Steps of viral life cycle (viral binds to host cell; lock and key fit between viral protein and host cell surface receptor; viral genome, via variety of mechanisms, enters cell; viral genome 'commandeers' its host, using host cells machinery to copy viral genome and synthesize viral proteins; DNA viruses usually use the host cell's DNA polymerase-such as reverse transcriptase).
Animal Viruses
[edit | edit source]Animal Viruses are very diverse with many modes of reproduction. Many animal viruses have outer membrane (viral envelopes). Typically there is a lipid bilayer (derived from the host cell plasma membrane) with virally encoded proteins protruding from it. These virally encoded proteins are important for binding and helps virus enter the cell.
Herpes virus envelop derive from nuclear envelope (herpes virus genome integrated into host cell DNA - as provirus). Usually the virus remains latent. But stress causes virus to become active. The provirus leaves genome and initiates viral production. Blisters form as a result.
DNA Viruses
[edit | edit source]There are two types of DNA viruses: double stranded DNA viruses and single stranded DNA viruses.
•Double stranded DNA viruses, such as the Small Pox virus, are able to use DNA polymerase to replicate itself, RNA polymerase to transcribe mRNA, wherein it will then use the host cell's ribosomes to produce proteins.
•Single stranded DNA viruses will have to use DNA polymerase twice to replicate its own genome (first synthesized strand will be a conjugated gene expression of the viral genome), but they can still use RNA polymerase to produce mRNA and the host cell's ribosomes to produce proteins.
RNA Viruses
[edit | edit source]There are three types of RNA viruses: double stranded RNA viruses, single stranded RNA viruses, and the retrovirus. Unlike DNA viruses, RNA viruses can use RNA replicase to replicate in a host.
•In a double stranded RNA viruses, each strand is considered to be viral mRNA. RNA replicase can be used to both duplicate the genome and produce viral proteins.
•There further exist two types of single stranded RNA viruses, positive and negative. In positive SS RNA viruses, the RNA is known as a "sense" strand, and acts as the mRNA. RNA replicase will make a complement of the mRNA, which can then act as a template to replicate it's genome. Positive SS RNA viruses also use RNA replicase to translate proteins. In negative SS RNA viruses, their genome is considered a "nonsense" strand, and is not the same as the mRNA strand. As a result, RNA replicase must make a template strand off of the "nonsense" strand and this can then be used to produce proteins. It also serves as a template to replicate new, "nonsense" RNA.
•Retroviruses are also single stranded RNA viruses, however, their genome is converted into double stranded DNA through viral reverse transcriptase. First, Reverse transcriptase will make a DNA/RNA hybrid strand using the original single stranded RNA genome. Then it will use the newly synthesized single stranded DNA template to make a double stranded DNA, and then integrate it into the host genome where it will be duplicated during the host cell's cell cycle.
Viroids and Prions
[edit | edit source]Viroids are small molecules of naked RNA that infect plants. Prions are infectious proteins (cause a number of degenerative brain disease). Brain diseases: mad cow diseases, scrapie in sheep, and Creutzfeldt-Jakob disease in humans. It may be a misfolded version of a normal protein, which redirects protein folding in infected cells. This generates more misfolded protein which can infect other cells.
Fighting Viruses
[edit | edit source]Host Cell Defenses
Viruses are often hard to see in cells because their "envelope" makes them look like parts of the host cell. However, if the cell sees double stranded RNA, it alerts the cell that something is wrong. Double stranded RNA is considered to be foreign, and once it is spotted, dicer enzyme will cleave it into smaller RNA pieces. RNA interference (RNAi) will come and destroy the small double stranded RNA pieces. Double stranded RNA will also signal transcription to stop, and increase the amount of ribonucleases, an enzyme that catalyzes the degradation of RNA. This will get rid of all the double stranded RNA as well. Once all the double stranded RNA is destroyed, transcription will continue as normal.
If a virus inhibits dicer and the cell can't eliminate the virus inside the cell, the infected host cell will be tagged for apoptosis.
An immune response through the Major Histocompatibility Complex I (MHC I) occurs as well. MHC I are involved in antigen presentation. Once the host cell cleaves the virus into smaller pieces, MHC I can present the antigen outside the cell, which can then be recognized by T cells and promote antibody production to help kill the infected cells.
Treatments
Viruses have structures that easily change conformation to adapt to new hosts and conditions, which explain how they elude drug treatments.[2] For example, there are new flu shots available every year to combat new strains of the flu virus, which arise from minor mutations that allow them to better exist in certain conditions.
Antibiotics do not work on viruses, instead, vaccines are synthesized invitro. Vaccines are made through purified viral proteins. The purified protein, lacking the virulent genomic information, can be cleaved into pieces and injected into a host. This way, the host will not risk the virus infecting its cells, rather, the body will recognize the cleaved pieces as foreign antigen from the virus and mount an immune response against it. This enables the immune system to have antibodies prepared and be ready in the event of a real infection.
Reverse transcriptase inhibitors (RTIs) are a type of antiretroviral drugs, that target RNA viruses such as HIV infections. RTI is used to inhibit the activity of reverse transcriptase which is a required enzyme that allows the reproduction of viruses. By inhibiting transcription, viral DNA is unable to be transcribed from RNA, which halts the viral infection cycle. However, with time HIV infected cells will eventually mutate into another form with a different kind of reverse transcriptase tag unaffected by antiretroviral drugs usually across a span of about five years.
Flu shots usually contain 3 different types of inactivated viruses, because researchers cannot always predict the exact type of virus that plays the biggest role in the seasonal flu. They hypothesize the protein structures of the 3 that have the highest probability and put those into the vaccine.
Resistance
Many antiviral agents are inhibitors that appear like the actual molecule that the virus' proteins want to bind to. By taking the place of the actual protein, the inhibitor is able to act like a "decoy" and block up the active site of the virus' protein and help stop the virus' life cycle.
Because the virus' protein's active site cannot easily be changed, the effectiveness of the inhibitor being used depends on how well it fits into the active site. Thus, oftentimes, the closer that the inhibitor's structure is to the actual molecule that usually fits into the active site, the more effective the inhibitor. It has been found through clinical tests that the closer the inhibitor's structure is to the actual substrate, the higher the barrier to resistance by the virus.
Viruses also have machinery that allows them to fuse with host cells. This fusion machinery has also become a primary target of drugs. Viruses such as HIV have shown that membrane fusion follows the formation of a six-helix bundle. This bundle is created through a pathway in which there is a trimeric coiled-coil intermediate which can associate with the target membrane by use of a "fusion peptide" at the N-terminal end.
Large problems with viral resistance have arisen. Higher than normal rates of viral resistance within a population can often be attributed to patient use of the antiviral agent. By not taking the proper dose or not using the drug for the prescribed duration of time helps lead to viral resistance. This is because without the proper dosage of the drug (i.e.: less taken than prescribed), not enough of the inhibitor is present to take the place of the actual substrate. Thus, strains of the virus that are only mildly resistant to the drug and would normally die under the prescribed conditions will end up surviving and passing on its resistance genes to its progeny. After a certain amount of time, this weak resistance is able to become strong enough after multiple mutations to fight off the antiviral agent, even under prescribed conditions. Through the selection process of non-resistant strains being removed from the genepool, more and more strains are becoming resistant.
A large problem with anti-viral resistance is the responsibility that everyone in a given population has to help prevent its spread. One person's negligence in not taking his or hers prescription properly can help the resistant strain spread - giving other people less of a fighting chance when they themselves become infected. Unfortunately, even with the utmost diligence in following a given prescription, viruses can still become resistant after a certain amount of time. So even though resistant viral strains are ultimately becoming more and more common no matter what we do, people should still take personal responsibility to wholeheartedly follow a given regiment to help slow the spread of this phenomenon.
Viral Benefits
[edit | edit source]There are a few benefits that viruses provide for humans.
•Viruses can help us to understand our cellular machinery.
•Viruses attack harmful insects and bacteria.
•Viruses also provide a hope for gene therapy. Gene therapy is the process of introducing foreign DNA into a cell. In theory, one can obtain an empty virus (one that's had its disease-causing genome removed), load it with a specific DNA, amplify the virus through PCR to have a specific protein that can target receptors on a specific cell, and have it inject the DNA into the cell. For example, cystic fibrosis occurs due to the lack of CFTR, so in theory, one can load an empty virus with CFTR DNA, tag it with something that's specific for lung receptors, and inject it into a host. However, in reality this is very difficult to do. Furthermore, viruses have a relatively high transfection rate. In other words, they are able to successfully transmit their genetic material into the host cell at a high rate and with less complications than more traditional methods that often cause unnecessary cell death.
Influenza
[edit | edit source]The influenza virus is a negative single stranded RNA virus. 2 types pose the greatest threats to humans: type A and type B. It has eight strands of RNA that encode for eleven different genes. These genes are then translated into many different proteins, but two major types of viral proteins are:
•Hemagglutinin (HA), 16 different antigens exist Hemagglutinin is found on the surface of the viral protein and binds to the sugars.
•Neuraminidase (NA), 9 different antigens exist Neuraminidase (sialidase) cleaves the oligosaccharide chains which releases the virus' progeny to infect other cells. This viral protein has been targeted for anti-Influenza treatment. Neuraminidase inhibitors including Zanamivir and Oseltamivir (2 anti-Influenza drugs) are different structural analogs of the substrate that attempt to fit into the virus' neuraminidase binding site in order to prevent it from attaching to the actual cellular receptor. 10 distinct forms of neuraminidase have been discovered - 9 of them called "N1, N2, N3, ..., N9" for Type A Influenza and "Type B neuraminidase" for Type B Influenza. Although Type A and B's neuraminidase catalytic machinery are essentially the same, differences in the amino acid sequence in other regions of the protein cause slightly different reactions of each towards various neuraminidase inhibitors.
Current research in neuraminidase inhibitors has found that many analogs of current inhibitors preferentially bind to Type A influenza over Type B, making them poor choices as inhibitors. These changes that make the inhibitor select preferentially for Type A include analogs of the carboxamide chain in Neu5Ac2en.
The N1, N4, and N8 subtypes of neuraminidase in Type A Influenza are special, relative to the other subtypes because they contain an additional subsite within the binding site. The amino acid sequence responsible for this has not yet been identified. Furthermore, no biological function has been found to be associated with this additional subsite.
HA is a viral protein that binds to the host cell for entry. It specifies which cells can be infected. NA is the viral protein that is responsible for viral release by cleaving the HA/host cell interaction.
Influenza strains consist of different combinations of HA and NA, as well as other viral proteins.
A growing concern among many is the lethality behind the different strains of influenza. For example, the Bird Flu, Swine Flu, and the common flu.
•Bird Flu (H5N1): This strand does not infect humans easily and is not easily spread. There have only been 400 cases in the last 5 years, however, there were 262 deaths, giving H5N1 ~65% mortality rate (11/2009). H5N1 targets the lungs, and while it is not something many have to be concerned with at the present time, it will grow to be a very severe problem if the strand mutates into a form where transmission can occur from human to human.
•Swine Flu (H1N1): This strand, unlike H5N1, infects humans easily and can be spread easily. There have been 14-34 million estimated cases, with around 2,500-6,000 deaths, giving H1N1 less than a 0.01% mortality rate (11/2009). This shows that H1N1 is not very virulent, which is very good for humans. However, if the strand ever becomes able to mutate into a virulent form, it would become a very severe problem.
•Common Flu: The common flu is constantly mutating each year, and there is an estimate of 25-50 million cases a year with around 30,000-40,000 deaths.
Resistance
[edit | edit source]When Neu5Ac2en analogs were cultured with neuraminidase inhibitor-resistant influenza viruses, off-target mutations in receptor-binding sites of hemagglutinin occurred. This revealed a balance between the efficiency of demolition by neuraminidase and hemagglutinin’s affinity for the receptor.
Experiments were done with the catalytic site variants of neuraminidase. There was a resistant virus chosen by one of the carboxamide analogs that had shown a mutation in one of the three argininyl residues, R292. That same mutation is a characteristic known to all neuraminidases. A loss of enzyme activity can be achieved by substitution to K292, but a large loss of binding potency to carboxamide analogs of Neu5Ac2en will also occur. E276 has been proven to change the conformation of carboxamides bound with R292K variant to allow the hydrophobic pocket to bind. The interaction of E276 and R224 creates the hydrophobic binding pocket, but the resistance comes from the R292K variant. The resistance is a hydrogen bond between E276 and K292 seen in the crystal structure of the unliganded R292K variant.
The less the inhibitor resembles the substrate the greater the loss of inhibitory potency directed at R292K N9 neuraminidase inhibitors including: Neu5Ac, Neu5Ac2en, 4-amino-Neu5Ac2en, zanamivir, Neu5Ac2en carboxamides, and oseltamivir carboxylate.This finding supports the rule that the more a drug similarly resembles the natural substrates/ligands, the more effectively they will be able to suppress a drug-resistant virus. A minimal decrease of inhibitory activity toward the mutant and biological activity of the mutant is required in order to suppress a drug-resistant virus. The drug-resistant virus must preserve its binding ability in order to process the natural substrates or the drug that resembles the substrates.
Even though these in-vitro experiments are useful and educational, drug therapy on infected patients can prove to be quite different from these studies. The different dosages and methods of intake can result in different levels of drug at the site of the infection. A good example of this is the comparison of zanamivir and oseltamivir.
Bacteriophage
[edit | edit source]Bacteriophages are viruses that infect the bacteria. A good example is the bacteriophage T2, which infects Escherichia coli. The T2 and T4 bacteriophages have a capsid with a needle like tail that can insert their own genome into the host cell, where it can instruct the reproduction progeny virions. The newly formed virions can then escape the host cell through a common phenomenon known as lysis. The location where the lysis occurred can be conspicuously observed through the formation of plaques, a clear spot surrounded by a plethora of bacterial cells. Plaques form by a single phage particle that lyse a host cell which then can infect nearby cells.
Bacteriophage life cycle
To begin its life cycle, the phage must find a host to attach to its surface. Cell surface receptor, a protein found on the surface of the host cell that is specific to the viral component, properly mediates the attachment and contact of the phage to the host.
The cell surface receptor plays an important role for the host cell, but the virus has mutated and evolved to take advantage these receptors. An example can be seen with the lambda phage. E. Coli has proteins called “lambda receptor proteins,” that allow the bacteria to acquire the sugar maltose for metabolism. However, the lambda phages have specific receptivity to the maltose porins in the outer membrane of E. Coli. Although this is detrimental for the host bacteria, natural selection has kept this porin for metabolic reasons.
With the insertion of the phage genome into the host cell, it directed the host to produce the progeny phages. Alfred Hershey and Margaret Chase In 1952 experimentally proved this when they have experimentally shown that when they transferred the DNA by a bacteriophage to a host cell, it led to the production of progeny phages. In 1950, Andre Lwoff and Antoinette Gutman observed that the phage genome can be integrated within the genome of the host bacteria. Most bacteriophages only insert their genome into the host through the cell envelope without the need for the whole capsid to penetrate the cell wall. T4 for example exhibit this kind of behavior. This virion has a neck tube that can contract and insert its DNA through the cell surface and into the host cell’s DNA.
Bacteriophage can go through two main cycles: The Lytic and Lysogeny cycle.
In a lytic cycle, the phage directs the immediate production of its progeny following the insertion of DNA. The process involves both the replicating the phage genome and the expression of the bacteriophage mRNA for the production of enzymes and capsid proteins. In some phages, like the T4, the host DNA is digested and increase the efficiency and of the bacteriophage production. After a plethora of progeny phage gets created, it proceeds to host cell lyses, which then releases the phages. Lysis is often referred to as a burst and the number of virus progenies released is called the burst size.
In the Lysogenic cycle, the phage inserts the DNA into the host cell but integrates its own genome into that of the host cell. Phage lambda, which has a linear genome of a double stranded DNA, reshapes the DNA to a circular shape upon entry into the host cell. The circular DNA can then integrate into the host genome by site-specific recombination of DNA. In this recombination, the recombinase enzyme aligns the Bacteriophage DNA with the host DNA so that the phosphodiester backbone links can be exchanged, which then leads to the integration.
The integration allows the phage genome to be replicated along with of the host cell as it replicates. The phage genome in the host DNA is called the prophage. Not only does Lysogeny integrate the phage genome into that of the hosts but also it can spontaneously generate a lytic burst of a phage. The prophage directs its own removal from the host genome by intramolecular process of site specific recombination with the two ends of the phage genome exchanging the phosphodiester backbone linkages once again. While the excised DNA exits the host genome, it circularizes and commences the lytic cycle, thus destroying the host cell and releasing the progeny phage.
Bateriophage can also go through a less prominent cycle called the Slow release cycle performed by the filamentous phages like the M13 phage. In this particular cycle, the phage particles replicate without the lysis of the host cell. The single stranded circular DNA of M13 serves as a template strand for the synthesis of a double stranded intermediate. Then this intermediate produces singe-stranded progeny genomes that get packaged by coating and supercoiling with the capsid proteins. These progeny phages force out through the host cell envelope without lysing the cell. The host cell continues to reproduce but more slowly because much of the resources are used to the production of the virus.
Proteins that bind DNA and subdue the transcription for the replication of the virus decide whether to go through the Lysis or Lysogeny. The transition from Lysogeny to Lysis can occur randomly but can also be affected by environmental factors such as UV light, which can damage the cell’s DNA. As for environmental cues, if a host cell’s growth is very strong, it’s more frequent to see the phage DNA inactive while an event that threatens the survival of the cell will initiate the lytic phage.
Virus transferring host genes
During the exit from the lysogeny phase, the phage can acquire the host genes and pass it onto another host cell in an event known as Transduction. Sometimes the whole phage genome can be permanently replaced by the host genome and packaged into the capsid, which would only be capable of transferring host DNA.
Lock and Key Mechanism
[edit | edit source]All viruses have a viral capsid. A viral capsid is the coat and the genome. However, only some viruses have envelopes. Envelopes are made up of lipids and they are essentially the same as the plasma membrane of the cell; they are similar because envelopes actually come from the host plasma membrane. Thus, the envelope has host and viral proteins that are gotten by exiting the host: viruses cannot do anything from inside the cell, so they have to get out to infect more cells. They do so by budding off and taking a piece of the host plasma membrane. The goal of a virus is to duplicate its genome and make protein; however, the real goal is to make more viruses, and the virus does so by infecting the host cell and working inside the host cell. First, the virus needs to figure out what sort of cells it can get into. The viral capsid and envelope proteins define the host/virus specificity: in the capsid there are specific proteins that will interact with specific proteins on the host cell membrane. This is the lock and key mechanism, where only certain viruses will recognize certain host cells. The host cell range is the types of cells that viruses can infect, and viruses have a limited host cell range: viruses can infect only specific cells and not all cells in the body, and the host cell range is define by the lock and key mechanism. The virus gets in the host cell by endocytosis (like the endosymbiont hypothesis) or by genome injection where only the genome is injected into the cell (the entire virus does not need to get in the cell since it only really needs the genome in). Both endocytosis and genome injection depend on the lock and key mechanism in order to decide which viruses can get in.
Lytic Life Cycle
[edit | edit source]In this cycle, the virus gets inside the host cell, duplicates its genome, makes protein, and then assembles new viruses by getting together the pieces it just produced. Eventually, the virus will cause the host cell to burst, which means that the lytic life cycle results in cell death and non-enveloped viruses. Everything in this cycle is done as quickly as possible, and the virus then moves on to other cells.

Lysogenic Life Cycle
[edit | edit source]This cycle does not result in any immediate killing of the cell. The virus enters the cell and actually inserts its genome into the host cell's genome. Then, when the host cell genome is duplicated, the viral genome gets duplicated along with it. Then, when something signals the virus to duplicate, the virus enters the lytic life cycle. The lysogenic life cycle includes the lytic life cycle.
Enveloped Virus
[edit | edit source]This type of virus can undergo either the lytic or lysogenic life cycle, but the end is different. After assembling, the viruses start budding out of the cell. They do not immediately kill off the cell but instead they take bits of the host cell plasma membrane. Enveloped viruses therefore contain many similar proteins and lipids to the host cells, which means that they are difficult to be detected by the immune system. Non-enveloped viruses, on the other hand, are easier to spot because they are seen as foreign. Also, some of the viral characteristics make them detectable as foreign: their double stranded RNA (our bodies do not have double stranded RNA) and viral proteins such as RNA replicase and reverse transcriptase (which are also foreign to the cell). Nonetheless, viruses are not as easily detectable as bacteria: while bacteria float around by themselves making them vulnerable to faster detection, viruses hide inside our body cells.
Genome packaging
[edit | edit source]Viruses use mainly two methods to package their genomes
- Building a capsid around viral genome
- Building a capsid first, then package genome into capsid; very often require the use of motor proteins
Packaging Initiation
[edit | edit source]Before packaging its own viral genome, there must be a process that can help viruses differentiate their own genome from its host genome; this process is known as initiation. There are several ways to initiate packaging:
- Unsegmented RNA
- Capsid has a binding site complementary to the specific sequence of the RNA or DNA
- Segmented RNA
- Segments have complementary sequence to other segments
- Capsid has more than one binding sites that recognizes the difference sequences on different segments
- DNA
- Double stranded DNA use a relatively different mechanism.
Examples of motor proteins and their mechanisms
[edit | edit source]Different viruses have different types of genomes(dsDNA, dsRNA, ssDNA, ssRNA)and also different motor proteins. Not surprisingly, their packaging mechanisms differ from one another.
P4 ATPase
[edit | edit source]- Found in dsRNA viruses such as Φ6 and Φ12
- multi-subunit
- Has many other functions besides packaging genomes
P4 ATPase is a hexameric molecule that has a central channel that is lined with loops and helices[9]. Some of these loops have phosphate-binding sites. ATP will bind to these sites, resulting in a change in conformation within the central channel. This change in conformation is believed to be responsible for RNA translocation. P4 ATPase will remain in the final virion even when packaging is complete.
DNA motor proteins
[edit | edit source]- relatively powerful; most powerful known molecular motor is T4
- generates up to 60 picnewtons of force
- packages DNA at 700bp/s
Motor proteins that are used to package DNA genomes have to be more powerful than typical RNA motor proteins because DNA genomes have high density, which causes high pressures in the capsid(60 atm). Unlike P4 ATPase, DNA motor proteins will dissociate after completion of packaging.
Rotatory motor mechanism
[edit | edit source]One of the earliest motor protein component that was studied was the gp10 portal protein of Φ29. It was embedded within the capsid and its structure was determined to be similar to a funnal; the smaller end facing out of the capsid while the wider end faced the interior of the capsid. Its central channel was lined with α-helices that had negative charges on them[10]. This allows for the easy transition of DNA molecules since they are negatively charged themselves. It was predicted that the portal will rotate using energy from ATP hydrolysis, hence transporting DNA into the capsid. Cryo-electron microscopy discovered some structures on the capsid that supported this theory; however florescence spectroscopy experiments did not detect any rotation of the portal. This mechanism was therefore deemed unlikely.
Also, the shotgun method (also known as shotgun cloning) is a method in cloning genomic DNA. It involves taking the DNA to be cloned and cutting it either using a restriction enzyme or randomly using a physical method to smash the DNA into small pieces. These fragments are then taken together and cloned into a vector. The original DNA can be either genomic DNA (whole genome shotgun cloning) or a clone such as a YAC (yeast artificial chromosome) that contains a large piece of genomic DNA needing to be split into fragments.
If the DNA needs to be in a certain cloning vector, but the vector can only carry small amounts of DNA, then the shotgun method can be used. More commonly, the method is used to generate small fragments of DNA for sequencing. DNA sequence can be generated at about 600 bases at a time, so if a DNA fragment of about 1100kb is cloned, then it can be sequenced in two steps, with 600 bases from each end, and a hundred base overlap. The sequencing can always be primed with known sequence from the vector and so any prior knowledge of the sequence that has been cloned is not necessary. This approach of shotgun cloning followed by DNA sequencing from both ends of the vector is called shotgun sequencing.
HIV(Human immunodeficiency virus)
[edit | edit source]The HIV Virus is a kind of retroviridae called lentivirus. It infects vital cells in the human immune system and cause AIDS (acquired immunodeficiency syndrome) which will reduce the human immune system progressively. The HIV virus is difficult to fend off due to a few of its defense mechanisms including: Carbohydrate masking and the variance of its conformation.The patient will have a high risk of having life-threatening infections and cancers. HIV, just like other kind of Lentivirius, is transmitted as single-stranded, positive sense, enveloped RNA viruses. Unlike other retroviruses, HIV is roughly spherical with a diameter of 120 nm. It concedes of two copies of RNA that is positive single-stranded. The RNA is tightly bonded to nucleocapsid protein and the enzymes need for the development of the virus. There are two types of HIV, 1) HIV-1 and 2) HIV-2. HIV-1 is the majority of the HIV infection in the world since it is toxic and easier to infect other. It causes a progressive decrease of the CD4+T cell count. HIV-2 is infected per exposure. The HIV virus integrated into the host cell and become latent and cannot be detected by the immune system. There are 4 major ways to transfer HIV virus, unsafe sex, contaminated needles, breast milk, and transmission from mother to baby.
Some people, even if exposed to HIV, don’t develop AIDS because scientists discovered that they carried a rare genetic variant, which has slightly different sequence of nucleotides, that protects people from getting AIDS. It is called CCR5. This rare gene is thought to be selected during evolution because it made people resistant to an organism unrelated to HIV.
The time between getting the HIV virus in a human system and actually getting the disease associated with it (AIDS) is a very important factor to look into. During this time period, the human immune system gets progressively weaker, since the immune system is compromised. In fact, some viruses that people come into contact with (even the common cold) actually behave as cofactors to the HIV virus. However, to act as a cofactor, the other virus must have certain characteristics. First off, the other virus should be able to infect the same cells that HIV infects. Secondly, the amount of cells that get infected at the same time must be large enough to change the normal mechanism of the HIV virus. Many viruses have been proposed as cofactors to the HIV virus, but have failed to meet the first criterion listed.
Also, HIV viruses generally like to infect CD4+ T-cells. Many of these cells reside in the lymph nodes, so the HIV virus can also generally be found in the lymph nodes. HIV viruses can generally be found anywhere in the body that CD4+ cells are abundant. This includes places such as the adenoids, macrophages, and tonsils as well.
Many efforts have been made to create an effective vaccine against the HIV virus. Before being able to create the vaccine, the methods of transmission must be examined. This virus can be transmitted in four different ways. The most obvious of these ways is sexual transmission. The other paths of infection are needle sharing in drug users, mother to baby transmission, and the use of infected blood or products thereof. A good number of vaccines have been proposed for this, and are now in clinical trials. However, there are abundant ethical and social issues regarding the use of such vaccines on human volunteers. One concern is the obvious threat to human lives if the virus in the vaccine concoction gets out of hand. Another safety issue is that producing mass amounts of the retrovirus is hazardous to both the people working in the lab and the general public.
HIV Immunity
APOBEC is a protein that has been evolutionary conserved and is used in animals for making diverse proteins from mRNA. APOBEC3G is in the same family of conserved proteins, but is solely found in human beings. It is said to have an important job in anti-viral immunity, especially against retroviruses such as HIV, which is currently being studied. This symmetric protein, with 2 homologous catalytic sequences, is known to interfere with the reverse transcription activity of HIV before it can be integrated into the host chromosome. Typically, without APOBEC3G, a tRNA called tRNA3Lys binds to the HIV-1 primer binding site to start the process of reverse transcription. But when APOBEC3G is present, it can stop the primer binding site, which then stops the reverse transcriptase from making the single stranded DNA and eventually double stranded DNA.
However, there is something called the Viral Infectivity Factor (Vif), a protein that is native to the HIV virus, that is being researched because of its counteracting effects to APOBEC3G. Vif is known to attack APOBEC3G and deactivate it. Going back to how APOBEC3G functions, in the absence of Vif, APOBEC3G can catalyze dC to dU mutations in the reverse strand script, causing multiple copying errors in the daughter script. These are missense and nonsense codons that end up being copied.
Source: APOBEC3G: a Double Agent in Defense Harold C. Smith Department of Biochemistry and Biophysics and the Center for RNA Biology at the University of Rochester, School of Medicine and Dentistry, Rochester, NY 14642 USA
Reference
[edit | edit source]1. Berg, Jeremy "Biochemistry, 6th Edition" 2007
2. "Microbiology: an evolving science" by Joan L. Slonczewski and John W. Foster.
3. "Do viral proteins possess unique biophysical features?" by Nobuhiko Tokuriki1, Christopher J. Oldfield, Vladimir N. Uversky,
Igor N. Berezovsky, and Dan S. Tawfik.
4. "Biology" by Neil A. Campbell and Jane B. Reece
5. Colman, Peter M. "New Antivirals and Drug Resistance"
- ^ Sun, Siyang, Venigalla B. Rao and Nathan Nelson. “Genome Packaging in Viruses” Current Opinion in Structural Biology 20 (2010): 114-120. Pubmed. Web. 19 Nov. 2010.
Sun, Siyang; Rao, Venigalla B.; Rossman, Michael G. (2010), "Genome packaging in viruses", Current Opinion in Structural Biology, doi:10.1016/j.sbi.2009.12.006, PMID 20060706 {{citation}}: Unknown parameter |acess date= ignored (|access-date= suggested) (help)
Colman, Peter (2009), "New Antivirals and Drug Resistance", Annual Review of Biochemistry, PMID 19254207 {{citation}}: Unknown parameter |acess date= ignored (|access-date= suggested) (help)
6. The New Genetics - U.S Department of Health and Human Services
7. Colman, Peter M., “New Antivirals and Drug Resistance”, The Walter and Eliza Institute of Medical Research, 10.1146/annurev.biochem.78.082207.084029, March 2009, p. 95-112
8. Lever, A.M.L. "The Molecular Biology of HIV/AIDS". John Wiley & Sons. University of Cambridge Clinical School, UK. 1996.
9. http://www.ncbi.nlm.nih.gov/pubmed/22482909
History
[edit | edit source]When Human Immunodeficiency Virus (HIV) first began showing up in 1981 in the United States, medical professionals originally determined it to be a rare form of cancer that infected homosexual men, known as Kaposi Sarcoma. Emergency rooms in New York suddenly had a strange influx of seemingly healthy young males coming in with flu-like symptoms and Pneumocystis Jiroveci Pneumonia (PCP), a fungal pneumonia that only affected individuals with HIV. (Normally, this fungal infection affects everyone, but since people without HIV have normal, healthy immune systems, they can prevent the infection from spreading. However, individuals with compromised immune systems, like those with HIV, cannot dispose of the fungus properly and therefore get sick.) Within the first year of the outbreak, out of the 1600 cases that were present in New York, almost 50% of the individuals died from it. This lead to a state of panic and medical experts hurried to find a cure. The Center for Disease Control was able to link the disease to affecting something in a person's blood.
The virus was finally discovered in 1984 at the Institut Pasteur in France, but in 1985, a U.S scientist finally determined the cause of the Acquired Immunodeficiency Syndrome (AIDS), a term pioneered by the CDC, was caused by the HIV virus. However, a cure couldn't be made quickly enough, so by 1987, six years after the first public case, there were over 71,000 confirmed cases of AIDS, of which there were more than a 50% casualty rate. Nowadays, as science and medicine have progressed, multiple drug regiments have been created to combat the progress of the virus, giving affected individuals a healthier lives and longer lifespans.

+== Structure ==
HIV structure consists of the outer coat, known as the viral envelope, that is made up of two layers of lipids derived from an infected individual. On the virus membrane there are two different proteins embedded within it. A protein called gp120 is a glycoprotein that allows the virus to attach itself to the white blood cells within the human body. In addition, a protein called gp41 is another glycoprotein that allows the virus to pass through a cell. These proteins are the main focus to anti-retroviral medications because they are crucial in the infection of a host cell.
Within the viral envelope contains all of the genetic information that is needed to infect a host cell. A viral core consisting of 2000 proteins protects the viral DNA along with all of the enzymes necessary for infection, including integrase and reverse transcriptase. In addition, the virus contains three structural genes that allow it to make more virus particles as well having six regulatory genes to control the virus's ability of infecting a cell; these genes include tat, rev, nef, vif, vpr, and vpu.

How HIV Infects a Cell
[edit | edit source]One of the unique properties of HIV is that it is a retrovirus; it carries its genetic material as RNA and creates viral DNA with the help of an enzyme called reverse transcriptase. It targets the T-cells in the human body because it contains genetics information in the form of DNA, which is crucial to HIV infection. However other cells, like macrophages, B-cells, and monocytes can be infected as well. Through many years of research, the method by which HIV infects a cell has been revealed. The virus begins its infection by first binding itself to the receptor proteins on a human cell, which enables the membranes to fuse and allows the virus to enter the cell. Once inside the cell, HIV has an enzyme known as reverse transcriptase that makes viral DNA from the RNA already present in the HIV core. Next, the DNA gets integrated into the human DNA via the enzyme integrase that was also present in the viral core. At this stage, the host cell becomes the site of replication for the virus and it creates viral RNA and long virus protein strands. The proteins and viral RNA are assembled into immature cells and eventually pinch off and leave the mature cell. HIV protease, another key enzyme in HIV reproduction, cleaves the immature strands out and splices together the mature proteins (similar to the function of spliceosome making mature proteins from introns and exons) to create mature virus particles, which are then free to infect other cells. This cycles just propagates and amplifies as the virus infects more and more cells.

How is HIV Spread
[edit | edit source]HIV thrives inside semen, vaginal fluid, blood, and other bodily fluids. It is spread via unprotected sex, sharing needles, and breast-feeding. HIV cannot be contracted through saliva, kissing, or other means of physical contact.
APOBEC3G: a Double Agent in Defense Against HIV
[edit | edit source]APOBEC3G (A3G) is an enzyme of the cytidine deaminases family. It is a host defense factor against the proliferation of HIV viruses under experimental conditions. HIV viral cells can only bind to host cells without the display of A3G on the surface. Some HIV viral cells contain Vif, Viral infectivity factor that degrades A3G before invading the. HIV viruses without Vif will less likely be penetrating a host cell displaying A3G enzymes on its membrane. However, it is also observed that A3G has the ability to aid in the mutation and proliferation of HIV viral DNA inside host cells. By mutating the sequencial HIV viral DNA, the new viral DNA can transcribe and mature without being noticed. A3G undergo two different mechanisms that lead to opposite effects. The deaminase-dependent mechanism leads to the proliferation of mutated HIV viral DNA, and the deaminase-independent mechanism, which prevent HIV viral DNA from translating and maturing.
A3G Deaminase-independent mechanism
[edit | edit source]A3G contains a C-terminus and an N-terminus. Attached to the C terminus is a zinc-dependent deaminase (ZDD fold). This ZDD fold is responsible for all the deaminase activity that A3G undergoes. A3G was found to have an innate ability to bind nonspecific RNA and single strand DNAs. This property is essential to the deaminase-independent antiviral mechanism that A3G has to potentially stop HIV viral proliferation. When both A3G and HIV viral DNA are introduced inside a host cell. First, A3G binds to the nucleic acids within a host cell to block the tRNA strands that deliver the matching amino acids that the HIV viral DNA requires to elongate. Without the amino acids, the HIV viral DNA cannot transcribe and translate into mature viral DNA, thus, the antiviral activity of A3G.
A3G deaminase-dependent mechanism
[edit | edit source]The A3G deaminase-dependent mechanism binds to HIV viral single stranded DNA and catalyzes dC to dU in the primary strand. This point mutation can cause the translation of a mutated HIV viral DNA. After translation, the second strand would contain dA instead of dG. Some mutated viral DNA can integrate into the host cell chromosomes, leading to the expression of viral protein with missense or nonsense substitutions. Viral replication would continue to produce both HIV DNAs leading to viral proliferation. These mechanistic pathways are actually observed in HIV infected patients, where numerous dG to dA mutations were identified in their cellular sequences.
References
[edit | edit source]"Basic Information about HIV and AIDS." Centers for Disease Control and Prevention. Centers for Disease Control and Prevention, 11 Apr. 2012. Web. 21 Nov. 2012. <http://www.cdc.gov/hiv/topics/basic/>.
Cichocki, Mark. "The History Of HIV." About.com AIDS / HIV. N.p., 7 June 2007. Web. 20 Nov. 2012. <http://aids.about.com/cs/aidsfactsheets/a/hivhis.htm>.
Cichocki, Mark. "PCP - Pneumocystis Jiroveci Pneumonia." About.com AIDS / HIV. N.p., 7 June 2007. Web. 20 Nov. 2012. <http://aids.about.com/cs/conditions/a/pcpguide.htm>
NIAID. "HIV/AIDS." Structure of HIV. N.p., n.d. Web. 21 Nov. 2012. <http://www.niaid.nih.gov/topics/hivaids/understanding/biology/Pages/structure.aspx>.
Structure-Based Drug Design: From the Computer to the Clinic." The Structures of Life. N.p., July 2007. Web. 20 Nov. 2012. <http://publications.nigms.nih.gov/structlife/structlife.pdf>.
Overview
[edit | edit source]All cells require energy for continual survival and operation. This energy comes from energy-containing compounds such as sugars, starch or lipids. The breakdown and interconversion of these energy-containing compounds in living organisms is a biochemical process coined Carbohydrate metabolism.
Humans
[edit | edit source]Carbohydrate metabolism is carried out by aerobic respiration where glucose and oxygen are metabolized releasing water and carbon dioxide. In cellular respiration metabolic reactions in order to convert the energy stored in the carbohydrate into ATP (adenosine triphosphate). ATP is created and is often referred to as "the molecular unit of currency" for intracellular energy transfer. ATP stores the now broken down energy and transports it to different areas of the cell when needed.
Carbohydrates are stored as polysaccharides consisting of longer polymers of glucose(monosaccharides) by glycosidic bonds. When energy is needed or to be stored, these polysaccharides are cleaved into their smaller monosaccharides units in preparation for catabolism. Carbohydrate catabolism is this breakdown of larger carbohydrates into smaller pieces in order to retrieve the energy within the bonds.
There are also other types of Carbohydrate metabolism such as glycolysis, anaerobic respiration, glycogenesis and more.
Glycolysis
[edit | edit source]Glycolysis metabolic pathway is used by most microorganisms such as yeast, bacteria, animals, and humans. Glycolysis means the dissolution of sugar. Glycolysis begins with a single molecule of glucose (C6H12O6) and ends with the production of pyruvic (CH3COCOO- + H+). The pathway is catabolic (producing energy by converting complex molecules into simpler ones). The energy produced during glycolysis comes from the degradation of glucose and if stored as a molecule called adenosine triphosphate (ATP). The six-carbon glucose is reduced to two molecules of the three-carbon pyruvic acid. ATP synthesis is said to be coupled to glycolysis because the glycolytic sequence is produced by utilizing two reactions. Even though glycolysis is the primary system for forming energy, some organisms do not require oxygen, such as organisms like yeast, aerobic organisms. Hundreds of biochemical reactions in our bodies require the participation of ATP as a source of energy. These organisms can only gain a small amount of energy needed to function from this process. Glycolysis occurs two major steps, the first step involves the conversion of sugar to, glucose-6-phosphate. The second step is the conversion of the glucose-6-phosphate to pyruvate. Then the products of glycolysis are further metabolized and completely break down glucose. In some microorganisms lactic acid is a final product produced from pyruvic acid. This process is called homolactic fermentation. In some bacteria and yeast lactic acid is not produced in abundant quantities, instead pyruvic acid is made into ethanol and carbon dioxide. This system is called alcoholic fermentation. In tissues some organisms glycolysis is an introduction to complex metabolic machinery. This system converts pyruvic acid to carbon dioxide and water by using oxygen. The most common type of glycolysis is the Embden-Meyerhof-Parnas (EMP pathway), which was discovered by Gustav Embden, Otto Meyerhof, and Jakub Karol Parnas.
Embden-Meyerhof-Parnas
[edit | edit source]The Embden-Meyerhof- Parnas (EMP pathway) is a sequence of chemical reactions that breaks down glucose and releases energy that is captured and stored as ATP. One molecule of glucose makes two molecules of pyruvate and two molecules of ATP. The pyruvate then enters into the tricarboxylic acid cycle if oxygen is present it is further fermented into lactic acid. If not enough oxygen is present in the cell pyruvate is fermented into ethanol. Overall, glycolysis produces ATP and act as building blocks for other synthesis of other cellular products.
Anaerobic Respiration
[edit | edit source]Anaerobic respiration is one way of respiration that uses electron acceptors and oxygen. In anaerobes, compounds such as nitrate (NO3), sulfur (S), and sulfate (SO42-) are used. For the electron transport chain to work, a final electron acceptor must be present to allow electrons to pass through the system. In aerobic organisms, the final electron acceptor is oxygen. Anaerobic respiration is mainly used by prokaryotes that live in environments that do not have a lot of oxygen. Anaerobic respiration is energetically less efficient than aerobic respiration. Many anaerobic organisms will die in in oxygen and therefore can only use anaerobic respiration.
Aerobic Respiration
[edit | edit source]Anaerobic respiration is one way of respiration that uses electron acceptors and oxygen. In anaerobes, compounds such as nitrate (NO3), sulfur (S), and sulfate (SO42- are used. For the electron transport chain to work, a final electron acceptor must be present to allow electrons to pass through the system. In aerobic organisms, the final electron acceptor is oxygen. Anaerobic respiration is mainly used by prokaryotes that live in environments that do not have a lot of oxygen. Anaerobic respiration is energetically less efficient than aerobic respiration. Many anaerobic organisms will die in in oxygen and therefore can only use anaerobic respiration.
Plants
[edit | edit source]Plants utilize many of the same metabolic reactions as Humans to metabolize Carbohydrates. However, the cell walls chemical and physical properties restricts enzyme attack. Cell walls contain polysaccharides mostly comprised of cellulose and hemicellulose that store energy for the plant cells. Because of the complexities of cell walls, the complete deconstruction of these polysaccharides is very difficult due to the restriction of enzymatic attack upon them. In turn, the biochemical process of recycling of the energy gathered from photosynthesis is relatively inefficient.

Cellulases and hemicellulases are very complex and intricate enzymes that are composed of molecular structures where catalytic modules rapidly increase and contribute to protein-carbohydrate or protein-protein interactions. Cellulosomes are cellulases and hemicellulases that have been synthesized by anaerobes and assemble in a multienzyme complex. It is still unclear as to how the formation of these enzyme complexes are carried out, but it is suspected that anaerobic environments impose selective pressures which drives the formation.
Cellulosomes breakdown and deconstruct plant polysaccharides more efficiently than traditional methods (enzymatic aerobic respiration, etc.) hindered by the cell wall complexities. For example, C. thremocellum utilizes cellulose very rapidly and thus requires a dubious supply of energy. Cellulosomes have a specific activity against cellulose that is 50-fold higher than the traditional energy breakdown method(cellulolysis which uses anaerobic bacteria that produces cellulases enzymes). It is proposed that this increase in efficiency is due to the multienzyme macromolecular complex that potentiates and increases synergistic interactions between the catalytic units and the enzyme-substrate target. Although this hypothesis is reasonable and plausible, the actual reasoning behind this phenomenon is still unclear because of the numerous variable factors associated with cellulose hydrolysis. [3].
References
[edit | edit source]- ↑ G Cooper, The Cell, American Society of Microbiology, p 72
- ↑ Stetten, DeWitt Jr. and Topper, Yale J. Seminars on Carbohydrate Metabolism: "The Metabolism of Carbohydrates, A Review", American Journal of Medicine. Bethesda, Maryland
- ↑ Fontes, C M, & Gilbert, H J. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annual review of biochemistry, 79, 655-81.
Overview
[edit | edit source]Glucose is a monosaccharide that fuels cells. Since a high concentration of glucose destroys the osmotic state of the cell, resulting in cell damage, hence glucose is not used for energy storage. Instead glycogen, a form of glucose that serves the purpose of energy storage, is used. Glycogen is a multibranched polysaccharide that can be broken down into glucose and used as a source of energy.
4 enzyme activities are needed for the breakdown of glycogen in providing 6-phosphate:
- Degradation of glycogen
- Remodeling of glycogen
- Remodeled glycogen as a substrate
- Conversion of product
Degradation of glycogen: Production of glucose 1-phosphate
[edit | edit source]Glycogen phosphorylase is the major enzyme in helping with the breakdown of glycogen. Through the addition of orthophosphate, denoted Pi, glycogen phosphorylase cleaves the substrate to form glucose 1-phosphate. Phosphorylase sequentially removes the residues from the nonreducing side the glycogen by having the Pi to cleave the glycosidic linkage between C1 carbon and the oxygen. As a result, glucose 1-phosphate from the cleavage along with the enzyme phosphoglucomutase converts to glucose 6-phosphate.
Remodeling of glycogen: Participation of pyridoxal phosphate
[edit | edit source]- A proton from Pi is transferred to the oxygen on C4 of the glycogen chain and at the same time Pi grabs a proton from the pyridoxal phosphate (PLP).
- An intermediate, carbonium ion, is formed
- Pi attacks the carbonium ion, leading to the formation of α-glucose 1-phosphate along with the addition of a proton back to the PLP
Remodeled glycogen as substrate: Aid from a debranching enzyme
[edit | edit source]Cleavage of α-1, 4 bond stops when phosphorylase reaches the fourth residue away from the development of branch. Two more enzymes, transferase and α-1, 6-glucosidase, are introduced to further facilitate the remodeling of glycogen. The transferase moves three residues from one branch to another. Then α-1,6 glycosidic bond is hydrolyzed by α-1,6 glucosidase, releasing a glucose. In general, the goal of transferase and α-1,6 glucosidase is to turn the branched molecule to into linear form.
Conversion of product: Production of glucose 6-phosphate
[edit | edit source]The enzyme phosphoglucomutase has a phosphorylated serine residue at its active site, and the phosphoryl group is later added to glucose 1-phosphate, more specifically, at C6 hydroxyl group, to yield glucose 1, 6-bisphosphate. At the same time, the C1 phosphoryl group of the substrate is added to the serine residue of phosphoglucomutase, ending up with glucose 6-phosphate and phosphoenzyme.
Reference
[edit | edit source]Berg, Jeremy "Biochemistry", Chapter 21 Glycogen Metabolism. 615-620. Seventh edition. Freeman and Company, 2010.
General Information
[edit | edit source]Fatty acids are key constituent of lipids. Because of the hydrophobic properties that lipids contain, they are able to form membranes within organisms. These lipids possess their Hydrophobicity because of their fatty acids. The overall structure of fatty acids is long hydrocarbon chains of various lengths and degrees of unsaturation terminated with carboxylic acid groups. Some fatty acids have double bonds, which changes the structure. It is said that a fatty acid will usually have an even number of carbons.
Nomenclature
[edit | edit source]The nomenclature of a fatty acid is derived from the name of its parent hydrocarbon by substituting the final e with an oic (i.e., the C18 saturated fatty acid is called octadecanoic acid because its parent hydrocarbon is called octadecane). To number the carbon atoms within a fatty acid, you start at the carboxyl terminus. The C2 and C3 carbon atoms are often referred to as α and β carbons, respectively. At the distal end of the chain, the methyl carbon atom is often referred to as the ω-carbon atom. To denote the position of a double bond in the structure, it is represented by Δ, followed by a superscript number. Alternatively, the double bond can also be presented by counting from the distal end of the chain, with the ω-carbon atom as number 1 in the chain. Fatty acids are referred to according to their carboxylate form because they are ionized at physiological pH.

Saturated Fatty Acid
[edit | edit source]
In nature, most fatty acids exist as straight-chain hydrocarbons that attach to a carboxylic acid with the most frequent and even number of carbon atoms. The chain-length range is from 2 to 80 but commonly from 12 up to 24. With a chain length from 2 to 6, they are called short-chain, from 8 to 10 they are called medium-chain, and 12 up to 24 called long-chain fatty acids. Fatty acids are structurally simple and even with their derivatives can be subdivided into well-defined families. Among straight-chain fatty acids, the simplest are referred to as saturated fatty acids. They have no unsaturated linkages in the carbon backbone and cannot be altered during hydrogenation or halogenation process. Saturated fatty acids tend to be solid at room temperature and their melting points increase with increasing chain length.
Saturated fatty acids are most commonly found in animals. The most common saturated fatty acids are Lauric Acid with the chemical composition CH3(CH2)10COOH, Palmitic Acid with the chemical composition CH3(CH2)14COOH, and Stearic Acid with the chemical composition CH3(CH2)16COOH.
Unsaturated Fatty Acid
[edit | edit source]When double bonds are present, fatty acids are said to be unsaturated, monounsaturated if only one double bond is present and polyenoic if they have two or more double bonds generally separated by a single methylene group in the carbon backbone. The configuration for double bond is almost always cis. Therefore, the bent structure is common for unsaturated fatty acids. Unsaturated fatty acids tend to be liquid at room temperature; their melting point increases with increasing chain length but decreases with degree of unsaturation. When a fatty acid is unsaturated and has a short chain length, it increases the fluidity. Most commonly, unsaturated fatty acids are from vegetable origin.
The most common unsaturated fatty acids are Oleic Acid with the chemical composition CH3(CH2)7CH=CH(CH2)7COOH, Linoleic Acid with the chemical composition CH3(CH2)4(CH=CHCH2)2(CH2)6COOH, α-Linolenic Acid with the chemical composition CH3CH2(CH=CHCH2)3(CH2)6COOH, and Arachidonic Acid with the chemical composition CH3(CH2)4(CH=CHCH2)4(CH2)2COOH.

Types of Fatty Acids
[edit | edit source]Monoene acids are fatty acids that contain one double bond. The most common are C-16, C18 and C-22. They also usually have a cis double bond and the double bond typically lies on C-9 for the most important and abundant fatty acids. Oleic acid is one of the most common monoene acids because it is widely distributed and produced. Oleic acid is used as the prototype for all of the monoene acids and also for the n-9 family of polyene acids. Oleic acid can be found in olive oil and several nut oils such as almonds, filberts, cashews, pistachios, pecans and macadamia nuts.
Polyene acids are fatty acids with more than one double bond. Polyene acids that have a methylene-interrupted pattern of unsaturation with 2-6 double bonds and cis configurations are the most important. The two major groups of these methylene-interrupted polyene acids are the n-6 acids based on linoleic acid and the n-3 acids based on alpha-linolenic acid. Linoleic acid is the most common polyene acid because it is used as a prototype for other polyene acids and it is found in most vegetable fats. Alpha-linolenic acid is an essential lipid in leaves, stems and roots.
Another type of fatty acid is the oxygenated fatty acids. The most common oxygenated acid has a hydroxyl, epoxy, or furanoid unit. The most important hydroxyl unit is ricinoleic acid because is the major acid in castor oil, which is used in cosmetics, as a lubricant before and after hydration, and as a drying oil after dehydration. Vernolic acid is the most well-known epoxy acid. It is found in seed oils. Furanoid acids are found at low concentrations in fish oils.
Trans Fats
[edit | edit source]Virtually any candy that we pick from the store, if we care to read the contents says partially hydrogenated oils. The oils used in them are fatty acids. Chemically speaking any long carbon chains with a carboxylic group are fatty acids. By that definition Acetic acid is the smallest fatty acid and naturally occurring fatty acids can be as long as 20 carbons.
Saturated fatty acids are those that have all single bonds except for the keto carbon of the carboxylic group. Unsaturated fatty acids are those that at least have one double bond between the carbons. If the fatty acid has only one double bond, it is referred to as monounsaturated. If it has more than one double bond, it is a polyunsaturated fatty acid.
Cis and trans fatty acids:
All the fatty acids that are found in human body are cis fatty acids, with the exception of retinoic acid (which is present in the eye). If meats or fish are left outside exposed to air, they eventually start to reek. This is partly due to the oxidation of single bonds in the fatty acids, which turns them rancid and is responsible for the bad odor. However if they are saturated fats without single bonds, they do not smell.
Fatty acids with cis double bonds are liquids, although hydrogenation can turn them into solids by turning them into saturated fatty acids. Cis fatty acids have "kinks" in them and hence do not pack well, so they remain in a liquid state at lower temperatures. Saturated fatty acids, however, have straight carbon chains that pack well, which enables them to solidify up to higher temperatures.
The methods of hydrogenation of fats were developed in early 1900s for the purpose of developing solid fats for making soaps. Later they were used to hydrogenate dietary fatty acids such as soybean oils because hydrogenated oils do not go rancid and smell. Slowly, they got into baked goods and candies. And now-a-days, it is difficult to find any packaged foods or snacks without Trans fats.
Hydrogenation removes double bonds and not covert cis fatty acids to Trans fatty acid which also contain double bonds.
This leads to the process of hydrogenation:
While there are many modification to it, the major process is to heat the unsaturated oils to above 200 0C, add powdered nickel (as catalyst) bubble hydrogen through it. The double bonds get saturated. However all the double bonds do not get saturated. And at that high temperature some of the double bonds seem to migrate to other carbons in the chain. Formation of trans configuration is more stable than cis. In that process at the newer position they become trans double bond. Cis configuration has more strain in it than trans. As it is seen on the packet ingredient list, they are written as “partially hydrogenated".
Why are partially hydrogenated or trans fats bad for health?
As mentioned above, our body mostly contains cis fatty acids. Whether our cells make them or they are from natural diet sources, they are all of cis configuration. Since all the natural fats are cis, the cellular enzymes have active sites that preferentially metabolize cis fatty acids. So over several years, trans fats accumulate in the body over those of cis form. Since all the natural fatty acids are cis, the enzymes that synthesize triglycerides and the enzymes that breakdown fats for energy, may not work efficiently. If they are not natural molecules, the cell’s enzymes can’t either break them or break them inefficiently. In addition as the trans fats accumulate in the body, as they are similar in structure to cis fats (to an extent) they mat act as competitive inhibitors to fatty acid metabolizing enzymes.
In addition, when natural cis fatty acids are incorporated into the cell membranes, as they have cis configuration, they do not pack very compact thus giving fluidity to the cell membrane. If membranes contain trans fats in them, the membrane fluidity will be affected. It is also likely that membrane receptor function will also be affected.
If the Trans fatty acids are incorporated into erythrocyte membranes, the membranes would be more rigid and erythrocytes would break as they travel through the microcapillaries.
Conclusions based on clinical studies
[edit | edit source]Women with high levels of trans fat in their bloodstreams had three times the risk of developing heart disease as women with the lowest levels of these kinds of fats. C-reactive protein (CRP) is made by the liver. Its levels in the blood are indicators of inflammation. A study of 700 nurses showed that those in the highest quartile of trans fat consumption had blood levels of CRP levels 73% higher than those in the lowest quartile. A 6 year study, monkeys fed with trans fats gained 7.2% body weight compared to 1.8% weight gain in monkeys fed with monounsaturated fats.
Biological Presence
[edit | edit source]Fatty acids in biological systems usually contain an even number of carbon atoms, typically between 14 and 24, although the 16- and 18-carbon fatty acids are the most abundant. Fatty acids typically contain an even number of carbon atoms because of the way in which fatty acids are biosynthesized. Animal fatty acids have hydrocarbon chains which are almost invariably un-branched. The alkyl chain may be saturated or it may contain one or more double bonds. In most unsaturated fatty acids, the double bonds are in the cis formation. The double bonds in polyunsaturated fatty acids, though, are generally separated by at least one methylene group. The chain length and degree of saturation give way to the properties that are found within the fatty acids and lipids. Unsaturated fatty acids have lower melting points than saturated fatty acids of the same length. Because double bonds cause the hydrocarbon chain to bend. Therefore, the fatty acids cannot compact tightly together, reducing the van der Waals interaction between the fatty acids. The melting point of fatty acids is also affected by chain length. The longer the hydrocarbon chain is, the higher the melting point. Short chain length and unsaturation enhance the fluidity of fatty acids and of their derivatives. Animals take advantage of this fatty acid property to maintain the fluidity of their cell membranes. When the weather turns cold, animals have an enzyme that converts saturated fatty acid to unsaturated fatty acid with one or more unsaturation. This prevents the membrane from getting frozen by lowering the melting point of the fatty acids in the membrane. Fatty acids can also form structures known as micelles in an aqueous solution. The structure is formed when the hydrocarbon tails form a hydrophobic center, while the polar heads form a hydrophilic shell outside the interior. The significance of micelles is that they act as emulsifiers, thus dissolving fat-soluble vitamins or other lipids that need to be absorbed.
Essential Fatty Acids
[edit | edit source]There are two fatty acids that the body cannot produce; all of the others can be derived from other molecules. Those two essential fatty acids are linoleic acid and alpha-linolenic acid. Luckily, these two can be found easily in most plant and animal oils. Other fatty acids such as omega-3 fatty acids can be produced by the body, but it is easier to get these from diet. Some sources are fish oils.
These fatty acids are used to help with essential body functions such as blood clotting, immune response, or blood pressure. They help make important fatty acids such as eicosanoids. Eicosanoids are important signaling molecules in the body. They are derived from 20-C chains derived from such molecules as the previously stated omega-3 fatty acids. Eicosanoids participate in activities such as relaying messages in the central nervous system or helping in the inflammatory response.
Other important Fatty Acids
[edit | edit source]Nutritional Significance
[edit | edit source]Fatty acids are significant in the nutrition of living organisms, because of the cell membrane's integral structure built up of fatty acids. Fats can be found in different quantities of various foods. While obesity is becoming a large issue in society today, the reviewing of types of fats and how they affect our bodies is a growing concern. Trans fatty acids are a large part of this concern. Trans fatty acids are fats found in foods such as some cookies, processed foods, crackers, candy, baked goods, fried foods, and other similar items. They are a concern to our health because studies have shown that diets high in trans fats increase the risk of various diseases including heart disease. The trans fats can be related to the levels of LDL cholesterol. Trans fats are found in ingredients labeled as shortening, and hydrogenated oil.
While fatty acids are an important part of living, they can be classified as "good" and "bad" fats. Bad fats are those that have negative effects on cholesterol levels. Bad fats are trans and saturated fatty acids. Good fats are those that have positive effects on cholesterol levels. Good fats consist of poly and monounsaturated fatty acids. Foods such as olive oil, soybean oil, and other vegetable derived oils are usually included. For example Butter consists of 29% Palmitic acid, 9% Stearic acid, 27% Oleic acid, 4% Linoleic acid, and 31% other. While olive oil consists of 6% Palmitic, 4% Stearic, 83% Oleic, and 7% linoleic fatty acids. The difference in the percents of saturated fatty acids between butter and olive oil is significant; butter has an overwhelming amount of saturated fatty acids compared to olive oil's composition being 90% unsaturated fatty acids. Beef as well is largely made up of saturated fatty acids, with 32% palmitic, 25% stearic, 38% oleic, 3% linoleic, and 2% other.
Isolation and Identification
[edit | edit source]The structure of a known acid can be defined by using gas chromatography, comparing the acid with an authentic sample or with compounds of related structure. However, if the fatty acid is completely unknown, spectroscopic procedures will be needed in order to provide more evidence. In order to determine the structure of a fatty acid you must know the chain length and the components of the structure such as branched or cyclic or other functional groups. You must also know the configuration, position, number and nature of the unsaturated centers and also the nature and position of the functional groups.
Thin-Layer Chromatography (TLC)
[edit | edit source]Used more for a qualitative comparison, TLC separates compounds with different polarities based off their attraction to the solvent (mobile phase)which moves up the TLC plate (stationary phase). The compounds have the choice to either react with either phase, which usually have different polar properties. The stationary phase is usually polar silica gel which ties up any of the more polar molecules, essentially slowing their movement up the plate, while the mobile phase is usually of lower polarity to move the less polar compounds father up the plate. Although thin-layer chromatography doesn't separate acids that differ only in chain length or degree of unsaturation, it is still useful in separating acids or esters with additional polar groups. If the silica layer is altered, then other separations can be achieved. One example of this is silver ion chromatography by inserting 5-20% of silver nitrate into the silica. This separates the acids based on the number of double bonds that they have.
High Pressure Liquid Chromatography (HPLC)
[edit | edit source]HPLC systems are essentially based off of the same principles of thin-layer chromatography, but have a more efficient separation process. By using a column with a thin internal diameter, coupled with a high pressured eluent to force the sample through the column, HPLC obtains a higher degree of separation than gravity powered chromatography. There are usually three main types of separation columns employed which are Gel Filtration (Size Exclusion), Ion-Exchange,and Affinity.
Gas Chromatography
[edit | edit source]Gas chromatography using capillary columns are now one of the most commonly used methods in separating methyl esters. By studying the elution out of the capillary column, chain length, degree of unsaturation, and the position of unsaturated centers can be identified.
Biosynthesis of Fatty Acids
[edit | edit source]Understanding the biosynthesis of fatty acids will give you information about the chemical pathway of the fatty acid, the enzymes that are involved in each step of the biosynthesis, the regulatory procedures, and where these reactions occur in the plant, animal, or micro-organism. The five major biosynthetic pathways are: the de novo synthesis of saturated acids from acetate, chain elongation, 9-desaturation to produce monoenes, desaturation in plant systems, and desaturation in animal systems.
Biosynthesis of a Butanoyl Group from Acetyl and Malonyl Building Blocks
[edit | edit source]Step 1: An acetyl group is transferred to the �-carbon atom of the malonyl group with evolution of carbon dioxide. Presumably decarboxylation gives an enol, which attacks the acetyl group.
Step 2: The ketone carbonyl of the acetoacetyl group is reduced to an alcohol function. This reduction requires NADPH as a coenzyme. (NADPH is the phosphate ester of NADH and reacts similarly to it.)
Step 3: Dehydration of the �-hydroxy acyl group.
Step 4: Reduction of the double bond of the �,�-unsaturated acyl group. This step requires NADPH as a coenzyme
De Novo Synthesis of Saturated Acids
[edit | edit source]All of the carbon atoms in fatty acids come from the two carbon atoms in acetate. One is half derived from the original methyl carbon while the other half is derived from the carboxyl carbon atom. Because fatty acids tend to be derived from C2 atoms explain why fatty acids almost always contain an even number of carbons. Malonate is more reactive than acetate and it is produced from acetate and carbon dioxide. Despite the involvement of malonate, it does not appear in the final product and all of the carbon atoms in the fatty acids come from acetate. This acetate-malonate pathway leads to three natural products, which depend on which synthetic pathway is followed. Acetate and malonate produce fatty acids from a reductive pathway, and also phenolic compounds by cyclization of polyacetate and isoprenoids by mevalonic acid. In the de novo pathway, acetate and malonate react in a condensation and reduction cycle, which produces the first C4 acid. The cycle is repeated and each time two more carbons are added until the fatty acid separates from the enzyme system using a hydrolase.
Chain Elongation
[edit | edit source]Chain elongation is very similar to the de novo synthetic route. Chain elongation is different in that the substrate is a preformed fatty acid that is saturated or unsaturated. The substrate reacts with acetyl, condenses, reduces, dehydrates, and reduces again. This produces another acid with another two carbon atoms, which are added at the carboxyl end of the molecule. This is the method by which many fatty acids are converted to long-chain acids such as palmitic acid to stearic acid.
Desaturation to monoene acids
[edit | edit source]Typically, unsaturated acids are produced by aerobic pathways. This occurs by inserting a double bond into a saturated acyl chain in the 9 position. To do this, you remove the pro-R hydrogen (stereospecifically and regiospecifically) from the C9 and C10 carbons to produce a cis alkene.
Desaturation to polyene acids
[edit | edit source]To desaturate further, you must insert more double bonds. In plants, an additional double bond is introduced between the existing double bond and the methyl group, creating a cis configuration. Plants can also introduce double bonds between the existing double bond and the carboxyl group; however this is much less common. Animals are not able to introduce double bonds on the methyl side of the n-9 double bond. Therefore, animals must derive the required linoleate and alpha-linolenate from plant-based dietary sources. Once these are derived, the fatty acids in animals can be desaturated and chain elongated.
Chemical Synthesis of Fatty Acids
[edit | edit source]A lot of the common fatty acids can be collected from natural sources and purchased from chemical suppliers. However, chemical synthesis of fatty acids may necessary if the acids are not easily collected from natural sources because there is no easily accessible source, if acids don’t naturally occur, or if acids are needed in their isotopic form. Saturated acids can be easily formed by chain extension of already available starting acids. This can also be applied to monoene and some other unsaturated acids.
Synthesis via acetylenic intermediates
[edit | edit source]Typically, though, the chemical synthesis of unsaturated acids involves the use of acetylenic intermediates, also known as the Wittig reaction. Acetylene can be alkylated once or twice. Also the triple bond in acetylene can be partially reduced to give cis- or trans-olfenic compounds. The reactivity of alkynes can be extended to give polyenes.
Synthesis by the Wittig reaction
[edit | edit source]In the Wittig reaction, an alkyl halide reacts with a base. This produces the ylid which is condensed using an aldehyde. The result can be either cis- or a trans-isomer. These can be distinguished by selecting certain reaction conditions. At low temperatures, high dilution and without the lithium ion, a cis-isomer is the result of the Wittig reaction. The final product can also be purified using a purification process such as silver ion chromatography.
Isotopically labeled acids
[edit | edit source]In order to produce an isotope of an acid, you must modify the above processes or incorporate into small molecules which are then synthesized. Isotopically labeled acids are essential for the study of reaction mechanisms and lipid biosynthesis and metabolism. The radioactivity of the products that come from the isotopically labeled acids can be analyzed. This is done using mass spectrometry or NMR spectrometry.
Utilization of Fatty Acids as Fuel
[edit | edit source]Peripheral tissues gain access to the lipid energy reserves stored in adipose tissue through three stages of processing. First, the lipids must be mobilized. In this process, triacylglycerols are degraded to fatty acids and glycerol, which are released from the adipose tissue and transported to the energy-requiring tissues. Second, at these tissues, the fatty acids must be activated and transported into mitochondria for degradation. Third, the fatty acids are broken down in a step-by-step fashion into acetyl CoA, which is then processed in the citric acid cycle.
Synthesis and Degradation
[edit | edit source]Although fatty acid synthesis is the reversal of the degradative pathway in regard to basic chemical reactions, the synthetic and degradative pathways are different mechanistically, showing that synthetic and degradative pathways are almost always distinct.
Important differences between the pathways
[edit | edit source]1. Synthesis takes place in the cytoplasm 2. Intermediates in fatty acid synthesis 3. The enzymes of fatty acid synthesis 4. The growing fatty acid chain is elongated 5. The reductant in fatty acid synthesis is NADPH 6. Elongation by the fatty acid synthase complex
References
[edit | edit source]Berg, Jeremy, Biochemistry, 6th Edition
Gunstone, Frank. Fatty Acid and Lipid Chemistry. Glasgow: Blackie Academic & Professional, 1996. 1-34.
http://www.elmhurst.edu/~chm/vchembook/551fattyacids.html
http://www.fda.gov/oc/initiatives/transfat/q_a.html
Chapters 12, 22, Biochemistry, Burg, Tymoczko and Stryer, 6th edition, W. H. Freeman and company
Chapter 19, Organic Chemistry, Vollhardt and Schore, 5th edition, W. H. Freeman and company
http://en.wikibooks.org/wiki/Cell_Biology/Membranes/Hydrophobicity
http://en.wikibooks.org/wiki/Structural_Biochemistry/pH
http://en.wikibooks.org/wiki/Analytical_Forensic_Pharmacology/Melting_Point
http://en.wikibooks.org/wiki/A-level_Chemistry/OCR_(Salters)/Isomerism
http://en.wikibooks.org/wiki/Organic_Chemistry/Introduction_to_reactions/Hydrogenation
http://en.wikibooks.org/wiki/Organic_Chemistry/Chirality/Configurations
http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme
http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Active_Site
http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Competitive_Inhibitor
http://en.wikibooks.org/wiki/Structural_Biochemistry/Chemical_Bonding/Van_der_Waals_interaction
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Micelles
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Fatty_Acids/Eicosanoids
http://en.wikibooks.org/wiki/Human_Physiology/The_Nervous_System
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Cholesterol
http://en.wikibooks.org/wiki/Structural_Biochemistry/Chromatography/Gas
http://en.wikibooks.org/wiki/Organic_Chemistry/Spectroscopy
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Fatty_Acids/Elongation_by_the_fatty_acid_synthase_complex Introduction Triacylglycerols, also known as triglycerides, are the simplest lipids formed by fatty acids. It is made up of three fatty acids ester linked to a single glycerol. Most triacylglycerols contain two or three different fatty acids. Triacylglycerols are nonpolar, hydrophobic, and insoluble in water. This is due to the ester linked bond between the polar hydroxyls of glycerol and the polar carboxylates of the fatty acids. Common triacylglycerols are vegetable oils, dairy products, and animal fat.[1]
Functions Triacylglycerols are stored as fat droplets in large amounts in vertebrate fat cells, and in plants as oils in the seeds. Triacylglycerol is a better stored energy source than polysaccharides because oxidation of triacylglycerols produces more than twice as much energy than the oxidation of carbohydrates. In addition, due to triacylglycerol's characteristic of being hydrophobic, it does not require hydration, thus it saves the organism the energy required to carry the extra water. [1]
Also, triacylglycerol located under the skin can function as insulation, in addition to being an energy source.[1]
- ↑ a b c Cox, Michael M. and Nelson, David L. Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. Print.
Cholesterol is a lipid with a unique structure consisting of four linked hydrocarbon rings forming the bulky steroid structure. There is a hydrocarbon tail linked to one end of the steroid and a hydroxyl group linked to the other end. The hydroxyl group is able to form hydrogen bonds with the nearby carbonyl oxygen of phospholipid and sphingolipid head groups. Cholesterol is known as a "sterol" because it is made out of alcohol and steroid. Cholesterol is present in most animal membranes with varying amounts but is absent in prokaryotes and intracellular membranes.
Cholesterol is also a key regulator of membrane fluidity in animals. It is able to insert itself into bilayers perpendicular to the membrane plane. The hydroxyl group forms hydrogen bonds with the carbonyl oxygen of a phospholipid head group while the hydrocarbon tail positions itself in the non-polar core of the bilayer. Since the structure of cholesterol differs from phospholipids, it disrupts the normal reactions between fatty acid chains. Cholesterol is also able to form lipid rafts when it forms specific complexes with certain phospholipids, which results in membranes that are less fluid and less subject to phase transitions. This also increases the permeability of the cell membrane to hydrogen and sodium ions.
Cholesterol is usually synthesized in animals and smaller cholesterol can be generated in plants. They are important in the composition of cell membranes and also steroid hormones.
The human liver makes 100% of the cholesterol needed for our body. There is no need to eat another animal to obtain any cholesterol. As the Institutes of Medicine of the National Academies has stated, "All tissues are capable of synthesizing enough cholesterol to meet their metabolic and structural needs. Consequently, there is no evidence for a biological requirement for dietary cholesterol." (Dietary DRI Reference Intakes. The Essential Guide to Nutrient Requirements. Institute of Medicine of the National Academies. The National Academies Press, Washington, DC.)
That statement highlights that the human body can eat a whole food plant-based diet (WFPB) and still make 100% of the cholesterol needed for the structural integrity of all its cells.

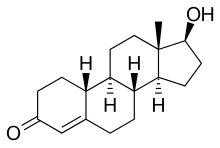

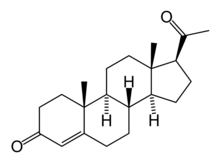

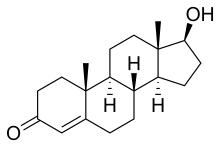

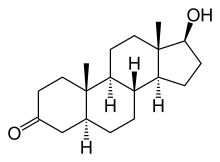

Cholesterol has played an important role in every cell in our body, especially abundant on the cell membrane, which functions as a communicator with other cells in our body. Small amount of cholesterol can also be found on the membrane of some organelles inside the cells, such as the mitochondrion and the endoplasmic reticulum.
Cholesterol is referred to as an amphipathic molecule, that it contains its hydrophilic and hydrophobic parts. The hydroxyl group (-OH) in cholesterol is aligned with the phosphate head of the phospholipid on the cell membrane, which the rest of the cholesterol goes with the fatty acid of the membrane. It is very important that cholesterol is present on all the cell membrane due to its properties keep the cell firm and avoid being overly fluid.
Another effect that cholesterol has in our body is that it promotes our learning ability and memory. It plays an important role in our nervous system. Good sleep can enhance learning ability and memory formation, and at the same time, a study showed that cholesterol synthesis increases during sleep. Not only cholesterol is abundant on the cell membranes, and also abundant in brain tissue in the nervous system. An important nerve cell, Myelin, covers nerve axons to help conduct the electrical impulses that make movement, sensation, thinking, learning, and remembering possible. The study showed that cholesterol was found to be the most important factor in the formation of synapses, which is greatly affecting our memory and learning ability.
Functions of Cholesterol
[edit | edit source]Cholesterol serves a variety of functions in the human body. This includes:
-The manufacture of steroids, or cortisone-like hormones, including vitamin D and the sex hormones testosterone, estrogen and cortisone. This in turn controls a myriad of bodily functions.
-Assisting the liver in the manufacture of bile acids, which is essential for digestion and absorption of fat-soluble vitamins such as vitamin A, D, E and K.
-Formation of the myelin sheath, a neuron consisting of fat-containing cells that insulate the axon from electrical activity. This ensures proper function of our brains by aiding route of electrical impulses. The absence of cholesterol might lead to loss of memory and difficulty in focusing.
-As a cell to interconnect "lipid molecules", which are needed to stabilize our cell membranes.
-As a source of energy
-Maintenance of our body temperature
-Protection of internal organs
- Modulation the fluidity of cell membranes
Health Significance
[edit | edit source]Cholesterol and triglycerides cannot dissolve in the blood. They have to be transported within the cells by carriers called lipoproteins. Low-density lipoprotein( LDL) and very low-density lipoprotein (VLDL) are known as the "bad" cholesterols while high-density lipoprotein (HDL) is known as "good" cholesterol.
Cholesterol Metabolism 1. LDL binds to a specific receptor, the LDL receptor (integral membrane protein) 2. Segment of the plasma membrane containing the LDL-LDL-receptor complex then invaginates and buds off from the membrane to form an internal vesicle. 3. The LDL separates from the receptor and is recycled back to the membrane in a separate vesicle. Vesicle containing LDL fuses with a lysosome leading to the degradation of the LDL and the release of cholesterol. This process can be beneficial because hormones and antibodies can transport proteins use this method. However, on the downside the pathway is also available to viruses and toxins as a means of entry into the cell.
During circulation, VLDL is converted in the bloodstream into LDL.
Hypercholesterolemia is a condition when there is an extremely high level of cholesterol in the body. Usually this means that there is a high concentration of LDL and low concentration of HDL. When too much LDL circulates the blood cell, it can build up on the inner walls of arteries that feed the heart and brain, and therefore, cause the clogging of the arteries. The health significance is that they are prone to cardiovascular diseases. If a clot forms and blocks the narrowed artery, a series of cardiovascular diseases such as hypertension, myocardial infarction, arteriosclerosis, angina pectoris, heart attack or stroke can result. High levels of cholesterol are also closely associated to diabetes.
HDL is known as "good" cholesterol in that it removes excess cholesterol in the arteries and transport it back to the liver for excretion or re-utilization, and thus preventing the arteries from clogging.
Hypocholesterolemia is a condition when there is an extremely low level of cholesterol in the body. This condition is usually rare, but if they do occur, it might be because of other illness that has caused the body to generate low or no cholesterols.
LDL cholesterol can be calculated with a rather rapid test based on the Friedewald equation, in which:
Total cholesterol = LDL cholesterol + HDL cholesterol + VLDL cholesterol
where VLDL cholesterol = triglycerides/5
One can rapidly and easily do a lipid profile by enzymatically measuring the important lipids—-total cholesterol, HDL cholesterol, and triglycerides. Dividing triglycerides by five gives the relatively unimportant, but hard to measure, VLDL cholesterol, which can in turn be used to calculate the important LDL cholesterol.
It should be noted that the equation does have certain limitations, and should not be used under circumstances:
•When chylomicrons are present.
•When triglyceride level exceeds 250.
•In patients with dysbetalipoproteinemia (type III hyperlipoproteinemia).
•Within about 12 hours after a meal, because triglyceride level can shoot up 20-30%
•Within 24 hours after the consumption of alcohol
Important numbers to know
| Desirable level | |||
|---|---|---|---|
| Total cholesterol | under 200 mg/dL | ||
| LDL, the bad cholesterol | under 100 mg/dL | ||
| HDL, the good cholesterol | over 40 mg/dL | ||
| Triglycerides | under 150 mg/dL | ||
Atherosclerosis- the result from contributor cholesterol
Atherosclerosis refers to the fatty deposits that block the arteries. Healthy arteries have smooth inner lining and blood can flow through them easily. However, if arteries have an infection it may result in rougher lining and cause inflammation. White blood cells go to the damaged arteries and begin to take up lipids, including cholesterol. Fatty acids start to grow at the affected area. This makes the artery become stiff and obstructs the blood flow. If unrecognized and untreated, atherosclerosis can lead to heart attack due to the death of cardiac muscle tissue. Drugs called statins which lower LDL (low-density lipoprotein(LDL), "bad cholesterol") can be used to treat atherosclerosis. Aspirin is also know to help against the recurrence of heart attacks.
Causes
[edit | edit source]High blood cholesterol can result from a variety of factors. For instance,
- Genetic factors
- Age
usually over age 45 in men and age 55 in women
- Gender
Before menopause, women tend to have lower total cholesterol levels than men of the same age. After menopause, women's LDL (bad) cholesterol levels tend to increase.
- Lack of physical activity
- Certain medications e.g., some diuretics, immunosuppressants, and corticosteroids
- Diseases like diabetes, hypothyroidism
- Cigarette smoking
- High dietary intake of cholesterol
- Obesity
Cholesterol content in food
[edit | edit source]The American Heart Association recommends a daily intake of less that 300 mg of cholesterol. Moreover, people with high LDL (bad) blood cholesterol levels or people who are taking cholesterol medication should limit themselves to a consumption of less than 200 mg of cholesterol per day.
Food sources and their corresponding cholesterol content can be found in the USDA National Nutrient Database. Top 20 of the list are:
| Cholesterol food source | Cholesterol content in mg/ 100 g food portion | ||
|---|---|---|---|
| Egg yolk, raw, fresh | 1234.9 | ||
| Chicken liver, all classes, cooked, simmered | 561.2 | ||
| Whole egg, cooked, fried | 456.5 | ||
| Chicken giblets, broilers or fryers,cooked, simmered | 442.1 | ||
| Whole egg, cooked, hard-boiled | 424.0 | ||
| Whole egg, raw, fresh | 424.0 | ||
| Beef liver, cooked, pan-fried | 381.2 | ||
| Whole egg, cooked, scrambled | 352.5 | ||
| Turkey giblets, all classes, cooked, simmered, some giblet fat | 289.0 | ||
| shrimp, mixed species, canned | 251.6 | ||
| Braunschweiger (a liver sausage), pork | 179.9 | ||
| shrimp, mixed species, cooked, breaded and fried | 177.8 | ||
| Hush puppies | 173.1 | ||
| English muffin, with egg, cheese, and Canadian bacon | 170.8 | ||
| Sponge Cake, prepared from recipe | 169.8 | ||
| Biscuit, with egg and sausage | 167.8 | ||
| Croissant, with egg, cheese, and bacon | 166.7 | ||
| Crab cakes, blue | 150.0 | ||
| Veal, leg (top round), separable lean and fat, cooked, braised | 134.1 | ||
| Eclairs, custard-filled with chocolate glaze, prepared from recipe | 127.0 | ||
Drugs used to lower blood cholesterol
[edit | edit source]Statins
[edit | edit source]This includes various drugs such as lovastatin, atorvastatin,simvastatin and cerivastatin etc. This class of drugs works by inhibiting the action of the 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase, an enzyme that catalyzes the conversion of HMG-CoA to mevalonate, an early and rate-limiting step in cholesterol biosynthesis.
Ezetimibe
[edit | edit source]This is a class of lipid-lowering compounds that selectively inhibits the intestinal absorption of cholesterol and related phytosterols.
Orlistat
[edit | edit source]Prevents cholesterol absorption by reducing the amount of pancreatic and gastrointestinal lipase
Fibrates
[edit | edit source]This includes fenofibrate and clofibrate. They stimulate the lipoprotein lipase, decrease LDL secretion and increase LDL receptor expression by binding to the peroxisome proliferator- activated receptor alpha.
Niacin
[edit | edit source]Niacin is a water-soluble B-complex vitamin as well as an antihyperlipidemic agent. It blocks the breakdown of fats, causing a decrease in free fatty acids in the blood and, consequently decreases secretion of VLDL and cholesterol by the liver. It also increases the level of HDL by lowering VLDL levels.
Anacetrapib
[edit | edit source]
Anacetrapib is an experimental drug that significantly raises good cholesterol while reducing bad cholesterol by almost half. It is a cholesterylester transfer protein (CETP) inhibitor being developed to treat hypercholesterolemia (elevated cholesterol levels) and prevent cardiovascular disease. Anacetrapid is an entirely new way of preventing heart attack and strokes mainly because not only it lowers LDL—the bad cholesterol, it also boosts HDL—the good cholesterol with no corresponding increases in blood pressure in any cohort. Elevated LDL and low levels of HDL are both risk factors for cardiovascular disease. Statins reduce LDL and lessen cardiovascular risk. Despite statin therapy, many patients still have a high risk of cardiovascular disease. High natural levels of HDL are associated with lower cardiovascular risk, which is why researchers were excited of the new drug’s effects, which helps keep fat particles attached to HDL, which carries them in the bloodstream to the liver to be disposed of. However, the drug will not be on the market anytime soon. It needs more testing.
References
[edit | edit source]Harvard Health Letter. November, 2004.
USDA National Nutrient Database for Standard Reference, Release 18 (2005).
http://www.cholesterol-and-health.com/Cholesterol-Cell-Membrane.html Structural Biochemistry/Lipids/Biosynthesis of Cholestrol/ Structural Biochemistry/Lipids/Lipoproteins/
Determining Membrane structure
[edit | edit source]Using a technique called freeze fracture electron microscopy scientist were able to discredit the membrane sandwich model by the discovery of transmembrane proteins. In this technique a membrane is frozen in liquid nitrogen and then shattered so the membrane splits between the leaflets of its bilayer exposing the interior of the lipid bilayer and its embedded proteins. Electron micrographs are then used revealing that the interior of the membrane is studded with globular membrane proteins and the outer surface has a relatively smooth appearance.
Bacteria Cell Walls
[edit | edit source]Bacteria cell walls are composed of Peptidoglycan, which create rigidity of the cell wall, determine cell shape, and help prevent osmotic lysis. The bacteria cell wall, like most lipid bilayers, is porous and semipermeable. Gram positive bacteria have a very thick peptidoglycan layer and therefore, when dyed during gram staining, retain the crystal violet dye. Gram negative bacteria have a thin peptidoglycan layer which results in an inability to retain the crystal violet dye during gram staining.
Peptidoglycan, also known as murein, is composed of alternating units of N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG). NAM is essentially a molecule of NAG onto which a lactyl group has been added to C3 via phosphoenol pyruvate; it is a signature molecule of bacteria. A pentapeptide chain is attached to the lactyl group during the NAM-NAG unit formation inside the bacteria. Generally, this pentapeptide chain consists of, in order: L-ala-D-Glu-mDAP/L-Lys-D-ala-D-ala. Once the NAG-NAM complex complex crosses the bacteria cell membrane into the periplasmic space, the terminal D-ala is cleaved off and the penultimate D-ala is cross linked to a DAP (forming a direct crosslink), or to a L-Lys (via a peptide interbridge) of another NAG-NAM complex. This crosslinkage is catalyzed by transpeptidases.
There are two cellular compartments that are involved in the biosynthesis of peptidoglycan. The formation, association, and assembly of soluble precursors onto a lipid carrier occurs in the cystoplasm. This generates Lipid II. This is a complex molecule that is thought to be translocated to the outer side of the cytoplamsimic membrane by integral membrane proteins of the shape, elongation, division, and sporulation family. The Lipid II's glycan chains are polymerized and the its stem peptides are cross linked. This is catalyzed by peptidoglycan syntheses which are penicillin-binding proteins, making this a target of B-lactam antibiotics. The biosynthesis of peptidoglycan involves over 10000 common reactions catalyzed in the cytoplasmic compartment.
The antibiotic penicillin works by competitively inhibiting the transpeptidase via the beta-lactam ring in the drug. Vancomycin also prevents the peptidoglycan layer from forming, but it works by binding to the terminal D-ala-D-ala complex, preventing transpeptidases from working. It typically is used as a last resort. Other drugs that work to inhibit peptidoglycan synthesis include phosphomycin, which inhibits NAM formation by competing with phosphoenol pyruvate, D-cycloserine, which inhibits enzymes that form the D-ala-D-ala unit inside the bacteria; and bacitracin, a drug that prevents bactoprenol from being recycled (bactoprenol is a carrier lipid, a homolog of dolichol, that moves the NAM-NAG complex across the cell membrane).
Additionally, bacteria have their own control mechanisms which inhibit peptidoglycan synthesis so that the organism can grow. These controls are classified as murein hydrolases, or autolysins, because they cleave various structures important for the murein layer. Hexosamidases cleave the beta-1,4 link between NAM and NAG, amidases cleave the peptide bond between the lactyl group and the pentapeptide chain on NAM, endopeptidases cleave various parts of the pentapeptide chain, and carboxylpeptidases cleave the D-ala carboxyl group, preventing crosslinking.
Penicillin-Based Proteins
[edit | edit source]Once Lipid II is on the periplasmic side of the membrane, it undergoes the polymerization of the glycan chains and the cross-linking of the stem peptides. While the Penicillin-binding proteins with high molecular mass carry out both of these reactions, penicillin-binding proteins with low molecular mass can exert carboxypeptidase or endopeptidase activities. These activities cleave peptide bonds within the stem peptide in order to regulate the level of peptidoglycan cross-linking. This insertion of the newly synthesized chains of peptidogylcan into the pre-existing layer is accompanied by the turnover of the old material produced by hydrolases (transglycosylases and endopeptidases). This sequence has proposed the idea that penicillin-based proteins are themselves associated with cell wall degrading enzymes within a comples that is located at the interface between the membrane and the periplasm.
Common Features that Biological Membranes Share
[edit | edit source]Composition of Membranes
[edit | edit source]The fluid mosaic model says that the three main components that make up membrane bilayers are lipids, proteins, and carbohydrates. In general, most membranes are composed of approximately 75% lipids, 20% proteins, and 5% carbohydrates.
Sheet-Like
[edit | edit source]Membranes are only two molecules thick and are sheetlike structures. They form closed boundaries and create different compartments. The thickness of most membranes are only between 6 nm and 10 nm.
Built of Lipids and Proteins
[edit | edit source]The mass ratio of lipids to proteins can range anywhere from 1∶4 to 4∶1. Although most membranes are made mostly of lipids and proteins some can have carbohydrates that are linked to lipids and proteins.
Composed of both hydrophilic and hydrophobic moieties (Amphipatic)
[edit | edit source]Membrane lipids are small molecules and they have both hydrophilic and hydrophobic moieties. The membrane lipids form closed bimolecular sheets. These lipid bilayers that are formed provide barrier to the flow of polar molecules. The hydrophilic head of the lipid molecule is on the outside surface of the cell membrane in addition to the inside surface of the membrane while the hydrophobic fatty acid long chains are on the inside of the lipid bilayer.
Proteins mediate functions
[edit | edit source]Different proteins mediate different function for the membrane. The membrane proteins are embedded in the bilayer and serve as pumps, channels, receptors, energy transducers, and enzymes.
Noncovalent
[edit | edit source]The proteins and lipids in the membrane are held together through noncovalent forces that work cooperatively.
Fluid
[edit | edit source]Membranes are fluid and lipid and protein molecules can freely diffuse into the plane of the membrane but do not rotate across the membrane. The membranes are two dimensional planes of oriented proteins and lipids.
Membranes are not static sheets of molecules locked in place but rather, they are held together by hydrophobic interactions which are much weaker than covalent bonds. As a result, most of the lipids and some of the proteins can diffuse laterally across the membrane. Adjacent phospholipids switch positions about 107 times per second where as molecules flip-flopping transversely across the member is rarely observed. The reason flip-flopping is rarely observed is because switching from one phospholipid bilayer to another requires the hydrophilic part of the molecule crossing the hydrophobic core of the membrane. Proteins on the other hand do drift laterally but at a much slower pace as their size, compared to lipids, is relatively large. Two factors increase the fluidity of membranes. The first factor is the presence of unsaturated hydrocarbon tails of phospholipids which have kinks that keep the molecules from packing closely together. The second factor is the presence of cholesterol which enhances fluidity at low temperatures by hindering solidification by disrupting the regular packing of phospholipids. What also enhances the fluidity of lipids is the lateral (fast) and flip flop (slow) exchange of place of lipid molecules between each other.
Asymmetric
[edit | edit source]The membrane does not match up and is asymmetric.
Asymmetry of membranes result for different functions needed by the cell as well as the molecular make up of each membrane. A membrane is a collage of different proteins embedded in the fluid matrix of the lipid bilayer. Phospholipids make up the majority of the membrane but proteins determine most of the membranes asymmetric properties as a result specification of membrane functionality. Different types of cells contain different sets of membrane proteins and the various membranes within each cell have a unique set of proteins necessary to carry out specific functions. An example of this is the sodium and potassium ion pump which uses ATP as a source of energy to pump ions in and out, but in order to do so, ATP must be inside of the cell to drive it.
There are two major populations of membrane proteins, integral proteins, which penetrate the hydrophobic core of the lipid bilayer, and peripheral proteins, which are no embedded in the lipid bilayer at all. Certain membranes contain distinct orientations of their integral and peripheral proteins, adding to the asymmetric character of the membranes. They have unique orientations because they are inserted into the membrane in an asymmetric manner after synthesis. Many of the integral proteins are trans membrane proteins, which completely span the membrane. The hydrophobic regions of an integral protein consist of one or more stretches of nonpolar amino acids usually coiled in alpha helices. The hydrophilic parts are exposed to the aqueous solutions on either side of the membrane. Peripheral proteins are loosely bound appendages on the surface of the membrane and are usually exposed to parts of integral proteins. Depending on the functions needed by the cell, whether it be transport, enzymatic activity, signal transduction, cell-cell recognition, intercellular, attach to the cytoskeleton and extracellular matrix, each membrane will look different. This asymmetry is preserved because membrane proteins do not rotate from one side to the other but "are always synthesized by the growth of preexisting membranes" (Berg, 345).
An example of membranes differing in their protein content is Myelin. Myelin is a membrane that serves as an electrical insulator around certain nerve fibers and has a low content of protein, 18%. Compared to the protein content of plasma membranes, 50%, the protein content is relatively low in Myelin. There are three major kinds of membrane lipids phospholipids, glycolipids, and cholesterol. The content of each in a membrane depends on the functions needed by the membrane and the characteristics of the cell.
Electrically Polarized
[edit | edit source]The inside of the membrane is negative at around −60mV.
Cell Growth and Forces that Shape It
[edit | edit source]Cell wall elongation and division of multi-protein complexes usually involves large macromolecues that are found on the outside of the cytoplasmic membrane. Penicillin-binding proteins, (PBPs) are intimately associated to the cytoplasmic cytoskeleton proteins. The bacterial cytoskeleton is a very important aspect of bacteria, and is the target of penicillin finghting antibiotics. Peptidoglycan is the macromolecule that determines the shape and maintenance of a bacteria cell by influencing the growth of the bacteria cell wall. There are two biochemical reactions that occur in the terminal stage of the peptidoglycan synthesis that is responsible for the lateral wall elongation and for septation. The two peptidoglycan-synthesizing systems are in competition with each other so that there is not any peptidoglycan synthesis for lateral wall elongation occurring during septation and not septation during cell wall elongation. The final shape of the bacteria is determined by these two cell wall synthesis interactions. A normal balance between the two reactions with result in bacteria being shaped like rods. An unusual prevalence of the site for elongation will lead to long rods or filaments. Any prevalence of septum formation will lead bacteria to the formation of coccobacilli or cocci. However, bacteria has a negative control that blocks septum formation when lateral wall elongation is not completed. This will help keep cocci from formation. Mecillinam is a Beta-lactam antibiotic that will specifically bind to penicillin, binding protein 2 (PBP 2). This will inhibit lateral wall elongation and transform rod shaped bacteria into cocci shaped bacteria. This can be extremely useful as the cocci shaped bacteria is unable to proceed to further division. However, it has been found that re-adding mecillinam to cocci that have not yet reshaped into rod shaped bacteria inhibits the peptidoglycan synthesis but does not interfere with the division occurring in the cells to which the antibiotic has been re-added even though these cells divide as cocci. This indicates that in order to release septum formation form the control that the lateral cell wall exerts on it, the cell wall does not need full elongation of the lateral wall. It has been found that cells with the removal of mecillinam, roughly one hour after removal, the cell division occurs. This suggests that the time needed for the septum to start and fully complete is approximately an hour. Because a normal rod will divide in half the time it takes for septum to complete, it is possible to deduce that the two septa are initiated at the same time in exponential cells growing under these conditions and are then stopped by the lateral wall elongation beginning. However, septum formation can be blocked by intracellular event such as DNA replication that postpone septum termination until such time as they are complete.
References
[edit | edit source]- Berg, Jeremy M. 2007. Biochemistry. Sixth Ed. New York: W.H. Freeman. 327
- Campbell, Neil A., Reece, Jane B. 2005 Biology Seventh Ed. Pearson Education, Inc.
Membrane proteins rely on their interaction with membrane lipids to uphold its structure and maintain its functions as a protein. For membrane proteins to purify and crystallize, it is essential for the membrane protein to be in the appropriate lipid environment. Lipids assist in crystallization and stabilize the protein and provide lattice contacts. Lipids can also help obtain membrane protein structures in a native conformation. Membrane protein structures contain bound lipid molecules. Biological membranes are important in life, providing permeable barriers for cells and their organelles. The interaction between membrane proteins and lipids facilitates basic processes such as respiration, photosynthesis, transport, signal transduction and motility. These basic processes require a diverse group of proteins, which are encoded by 20-30% of an organism’s annotated genes. There exist a great number of membrane lipids. Specifically, eukaryotic cells have a very complex collection of lipids that rely on many of the cell’s resources for its synthesis. Interactions between proteins and lipids can be very specific. Specific types of lipids cane make a structure stable, provide control in insertion and folding processes, and help to assemble multisubunit complexes or supercomplexes, and most importantly, can significantly affect a membrane protein’s functions. Protein and lipid interactions are not sufficiently tight, meaning that lipids are retained during membrane protein purification. Since cellular membranes are fluid arrangements of lipids, some lipids affect interesting changes to membrane due to their characteristics. Glycosphigolipids and cholesterol tend to form small islands within the membranes, called lipid rafts, due to their physical properties. Some proteins also tend to cluster in lipid raft, while others avoid being in lipid rafts. However, the existence of lipid rafts in cells seems to be transitory. Recent progress in determining membrane protein structure has brought attention to the importance of maintaining a favorable lipid environment so proteins to crystallize and purify successfully. Lipids assist in crystallization by stabilizing the protein fold and the relationships between subunits or monomers. The lipid content in protein-lipid detergent complexes can be altered by adjusting solubilisation and purification protocols, also by adding native or non-native lipids.
There are three type of membrane lipids: 1. Phospholipids: major class of membrane lipids. 2. glycolipids. 3. Cholesterols. Membrane lipids were started with eukaryotes and bacteria.
Types of Membrane Lipids
[edit | edit source]Lipids are often used as membrane constituents. The three major classes that membrane lipids are divided into are phospholipids, glycolipids, and cholesterol. Lipids are found in eukaryotes and bacteria. Although the lipids in archaea have many features that are related to the membrane formation that is similar with lipids of other organisms, they are still distinct from one another. The membranes of archaea differ in composition in three major ways. Firstly, the nonpolar chains are joined to a glycerol backbone by ether instead of esters, allowing for more resistance to hydrolysis. Second, the alkyl chains are not linear, but branched and make them more resistant to oxidation. The ability of archaeal lipids to resist hydrolysis and oxidation help these types of organisms to withstand the extreme conditions of high temperature, low pH, or high salt concentration. Lastly, the stereochemistry of the central glycerol is inverted. Membrane lipids have an extensive repertoire, but they possess a critical common structural theme in which they are amphipathic molecules, meaning they contain both a hydrophilic and hydrophobic moiety.
Although each membrane lipid is distinct there are several attributes that are universal. Membrane lipids are all closed bodies or boundaries separating substituent parts of the cell. The thickness of membranes is usually between 60 and 100 angstroms. These bodies are constructed from non-covalent assemblies. Their polar heads align with each other and their non-polar hydrocarbon tails align as well. The resulting stability is credited to hydrophobic interaction which proves to be quite stable due to the length of their hydrocarbon tails. The resulting structure is termed a lipid bilayer which mediates molecules that enter or leave the cell. Sugars are also a key structural component in cells by attaching themselves to lipids forming glycolipids which are used for identification of the cell by other molecules. Signaling molecules read the sugar patterns in glycolipids which then the glycolipids either allow or refuses entry of the cell. This process helps tissue and organ growth within the body. Phospholipids are another important component to such processes.
There are several other universal characteristics of membrane lipids such as they are electrically polarized attributing a negative charge within the membrane. This plays a role in many functions of the cell such as the transport of energy and the conversion of energy. Also, lipid membranes are asymmetric and fluid structures. Biological membranes lack symmetry elements and will diffuse without stable conditions. Lastly, lipid membranes have unique functions. These specific functions are due to membrane proteins
Phospholipids
[edit | edit source]
The major class of membrane lipids are the phospholipids. They are abundant in all biological membranes. Phospholipds are made from four components: one or more fatty acids, a platform to which the fatty acids are attached, a phosphate, and an alcohol attached to the phosphate. The fatty acid portion provides the hydrophobic barrier found in lipids, where as the rest of the molecule has a hydrophilic property, enabling interaction with the aqueous environment. Phosopholipds are built upon a foundation of glycerol, a three-carbon alcohol, or sphingosine. Phospholipids which are derived from glycerol are also known as phosphoglycerides, which consist of a glycerol backbone where two fatty acid chains and a phosphorylated alcohol are attached. The major phosphoglycerides come from phosphatidate through the formation of an ester bond between the phosphate group of phosphatidate and the hydroxyl group of one of several alcohols. Sphingomyelin is a phospholipid found in membranes that is not derived from glycerol. The backbone in sphingomyelin, however, is sphingosine, which is an amino alcohol which contains a long, unsaturated hydrocarbon chain.
Phospholipids have a very important property of separating compartments. For example, in evolution, it was very important in dealing with the RNA World Hypothesis. If RNA did not have a phospholipid bilayer, they would not have been able to contain all their chemical and mechanistic reactions in a certain space, and it would have been very hard for these RNA to survive without being disrupted by other arbitrary, outside reactions. Thus, phospholipid bilayers played a significant role in the production and survival of these RNA molecules.
Phospholipids have many unique functions. For example, they can function as a reservoir of intracellular protein messengers such as phosphoinositol biphosphate. Phosphoinositol biphosphate is one of the most important secondary messengers in the cell signaling pathway of human. Also, they anchor proteins to cells. This many determines the specific function of that lipid membrane. Phospholipids have a number of other functions making them the most abundant membrane lipid. These include energy storage, cellular shape, and it is a source of acetylcholine. Acetylcholine is a commonly occurring neurotransmitter found in both the peripheral nervous system (PNS) and the central nervous system (CNS).

Glycolipids
[edit | edit source]As the name implies, glycolipids are simply sugar-containing lipids. Glycolipds are typically composed of short, branched chains with less than 15 sugar units. The glycolipids in animal cells come from sphingosine, similar to those in sphingomyelin. As in sphingomyelin, the amino group of the sphingosine backbone is acylated by a fatty acid. Glycolipids have a unit that is linked to the primary hydroxyl group of the sphingosine backbone, which differentiates it from sphingomyelin. In glycolipids, one or more sugars are attached to this group. Glycolipids are arranged in an asymmetric manner with the sugar residues always on the extra cellular side of the membrane. The simplest glycolipid is called cerebroside which contains a single sugar residue. The sugar could be either glucose or galactose. Complex glycolipids, for example gangliosides, contain a branched chain of as many as seven sugar residues.
Glycolipids serve several important functions within the cell. These include shape, specific function, fuel storage, and a number of cellular tasks. For example, glycolipids flank from the extra cellular side of the membrane and serve as a marker for cellular recognition. The information many times tells a cell whether to grow, divide, or do nothing. Likewise, these markers can also be the antigens that correspond to blood type. Other molecules that serve as antigens to blood type are glycoproteins.
Cholesterol
[edit | edit source]Cholesterol is a lipid with a structure quite different from that of phospholipids. It is a steroid built from four linked hydrocarbon rings. A hydrocarbon tail is at one end of the steroid while a hydroxyl group is attached to the other end. The orientation of the molecule in a membrane is parallel to the fatty acid chains of the phospholipids, allowing the hydroxyl group to interact with the head group of phospholipids within the proximity. Cholesterol is absent from prokaryotes, but found in varying degrees in all animal membranes. Almost 25% of membrane lipids in certain nerve cells is made of cholesterol. Furthermore, cholesterol is also present in hormones found in the body. However, cholesterol is essentially non-existent from some intracellular membranes. Cholesterol contains four cycloalkane rings and OH group attaches at one end. Cholesterol is functioned as cell signaling and stays outside of the membrane. Cholesterol although it has a negative reputation has a number of important functions. There are two types of cholesterol HDL and LDL known as “good” and “bad” cholesterol, respectively. Low density lipoproteins (LDL) are known as bad because it tends to thicken the walls of the heart and arteries with plaque. This can lead to heart attacks, strokes, and a number of other health concerns. High density lipoprotein (HDL) is known as good cholesterol because it seems to do the opposite. It is believed that HDL cholesterol actually takes cholesterol from the heart back to the liver where it can be broken down and passed through the body. Cholesterol has a number of other functions important to the field of biochemistry. It is used to build membrane, supply the body with fat, and it also synthesizes hormones necessary for bodily function. Although it is necessary to find this membrane lipid in much lower quantities than phospholipids it serves several significant functions.

The sterol cholesterol is a major constituent of animal plasma membranes but is absent from prokaryotes. The fused ring system of cholesterol means that it is more rigid than other membrane lipids.As well as being an important component of membranes, cholesterol is the metabolic precursor of the steroid hormones. Plants contain little cholesterol but have instead a number of other sterols, mainly stigmasterol and beta-sitosterol which differ from cholesterol only in their aliphatic side-chains.
Membrane Lipids
[edit | edit source]Lipid Vesicles
[edit | edit source]Lipid vesicles, also known as liposomes, are vesicles that are essentially aqueous vesicles that are surrounded by a circular phospholipid bilayer. Like the other phospholipid structures, they have the hydrocarbon/hydrophobic tails facing inward, away from the aqueous solution, and the hydrophilic heads facing towards the aqueous solution. These vesicles are structures that form enclosed compartments of ions and solutes, and can be utilized to study the permeability of certain membranes, or to transfer these ions or solutes to certain cells found elsewhere.
Liposomes, with a diameter of roughly 500 A, are formed by dispelling a suitable lipid in an aqueous environment, and then sonicating the solution with high-frequency sound waves, for example, which aid these molecules to form a dispersed set of closed vesicles that are all nearly identical in size. Larger vesicles can be formed by placing a phospholipid in a solution containing both organic and aqueous solvents and then slowly evaporate the organic solvent. When this occurs with ions and solutes in the solution as well, that is when these compartments can entrap these ions and solutes in their lipid vesicles. The vesicles that contain the ions or solutes can then be separated by gel-filtration chromatography or dialysis. The permeability of a molecule can be obtained using this idea and then measuring the rate at which the molecule moves from the inner compartment of the vesicle to the outer solution.
The formation of liposomes is extremely useful in drug transport and delivery. Since drugs cannot readily diffuse through cell membranes, by creating liposomes in a solution containing a specific drug, the drug can enter into the cell by fusion of the liposome bilayer with the cell bilayer, delivering the liposome's contents. This technique can also be used to deliver DNA.
Liposomes as vesicles can serve various clinical uses. Injecting liposomes containing medicine or DNA (for gene therapy) into patients is a possible method of drug delivery. The liposomes fuse with other cells' membranes and therefore combine their contents with that of the patient's cell. This method of drug delivery is less toxic than direct exposure because the liposomes carry the drug directly to cells without any unnecessary intermediate steps. Long-circulating liposomes are more concentrated in regions of high blood circulation(like tumors and inflamed areas), and the selective fusion exhibited by liposomes allows them to be able to target specific types of cells. This is a very useful tool in designing carefully-controlled drug delivery techniques.
Lipid Bilayer
[edit | edit source]Because of the hydrophobic interactions among several phospholipids and glycolipids, a certain structure called the lipid bilayer or bimolecular sheet is favored. As mentioned earlier, phospholipids and glycolipids have both hydrophilic and hydrophobic moieties; thus, when several phospholipids or glycolipids come together in an aqueous solution, the hydrophobic tails interact with each other to form a hydrophobic center, while the hydrophilic heads interact with each other forming a hydrophilic coating on each side of the bilayer.
The cellular bilayer is not a rigid and clearly defined structure separating the intracellular and extracellular environments. Realistically, there is significant fluid motion of individual lipid headgroups and their aliphatic chains within the plane of the bilayer. This is due to the low torsional angle barriers within the structures of the lipids and to the steric hindrances that result from the cis-double bounds of some aliphatic chains.[1]
Due to the large variety lipids that can exist in a single cell, regions of cellular membranes will comprise of heterogeneous mixtures of lipids and membrane proteins.[2] The unique interactions within these heterogeneous regions are the bases of the function of the cellular membrane.

This lipid bilayer formation is spontaneous because of the hydrophobic interactions and energetically favorably structure. Other intermolecular forces such as Van der Waals, which hold the hydrophobic tails together, and hydrogen bonding, which bind the hydrophilic heads with water, help stabilize the lipid bilayer structure. Because of these interactions, the lipid bilayer inherits unique properties. The lipid bilayer have "extensive" properties, and can enclose and form compartments. Lastly, they can also recover quickly if there is a hole in the lipid bilayer, due to energetic reasons. However, phospholipids and glycolipids do not form micelles like fatty acids do because phospholipids and glycolipids have two fatty acids chains and are too big to form the interior of the micelle.
In aqueous solution, amphipathic molecules will orientate themselves in such a way as to prevent the hydrophobic region coming into contact with water molecules. In the case of those fatty acid salts which contain only one fatty acid chain (such as sodium palmitate, a constituent of soap), the molecules form a spherical micellar structure (diameter usually <20 nm) in which the hydrophobic fatty acid chains are hidden inside the micelle and the hydrophilic headgroups interact with the surrounding water molecules. Because the two fatty acid chains of phospholipids are too bulky to fit into the interior of a micelle, the favored structure for most phospholipids in aqueous solution is a two-dimensional bimolecular sheet or lipid bilayer.
Asymmetry of Lipid Bilayer
[edit | edit source]Plasma membranes are structurally and functionally asymmetric. There are different compositions and ratios of phospholipids inside and outside of lipid bilayer. In the plasma membrane, phosphatidylcholine and sphingomyelin are found in the outer monolayer (outer leaflet) but phosphatidylserine, phosphatidylethanolamine and the phosphatidylinositols are found in the inner monolayer (inner leaflet). Cholesterols are found in both leaflets.

The distribution of lipids between the lipid bilayer often changes to induce biological effect. A good example is that platelet moves phosphatidylserine to the outer leaflet to enable the ability of blood clotting. Phosphatidylserine is also an important signal for programmed cell death. In some cells, the presence of phosphatidylserine on the outer surface of cell membrane signals the immune system to destroy the marked cell. The asymmetry property of cell membrane enables cell to have multiple biological functions.
Membrane proteins are also inserted into the membrane in an asymmetric way.
In a lipid membrane, there are proteins embedded in them. The mass ratio of the lipid molecules and the proteins that are embedded in them ranges from 1:4 to 4:1. There are two types of proteins in lipid bilayer: integral and peripheral membrane proteins. Integral membrane proteins traverse the lipid bilayer meaning they interact extensively with the hydrophobic region (hydrocarbon region) of the lipid bilayer. Peripheral membrane proteins are usually attached to surfaces of integral proteins; therefore, they are on both faces of lipid bilayer. Peripheral membrane proteins interact with the hydrophilic polar head groups of the lipid molecule. Some peripheral membrane proteins can only be found outside the membrane or inside the membrane. This contributes the asymmetric property of membrane.

Protein Functions in Membrane
[edit | edit source]Proteins are responsible for the active transport of ions and larger molecules in and out of the cell membrane. They are classified as either integral or peripheral. Both forms of the protein are embedded in the lipid bilayer thru hydrophobic interactions. A typical integral protein spans the width of the lipid bilayer. They usually consist of many alpha helices, with their hydrophobic parts pointing outwards, into the membrane's hydrophobic sea of hydrocarbon tails. Alpha helices are non-polar and uncharged. This can be used to identify likely membrane - spanning regions. Free - energy can be estimated when a helical segment is transferred from the interior of the membrane to water. Each amino acid residue has a specific free energy change thus allowing us to know what the alpha - helices is made of. Free - energy chance for each window can be plotted against the first amino acid to create a hydropathy plot. Peaks of greater than 84kJ/mol have the potential to be transmembrane helices. When integral proteins are made of beta pleated sheets, they typically fold into a cylinder, once again, with the hydrophobic ends pointing out and the hydrophilic ends pointing in. These are called channel proteins and they usually work well as a form of ion transport because the hydrophilic core will allow charged particles to go thru. A peripheral protein is bound to the membrane by electrostatic and hydrogen-bond interactions with the head groups of the lipids. These interactions can be interrupted by the change of salt concentration and pH.
Built from the concept of integral and peripheral proteins, one biological significance comes from the structure of an archaeic protein called, bacteriorhodopsin. Bacteriorhodopsin consists of seven alpha helices that have a majority of nonpolar amino acids and few charged amino acids. The protein is a integral protein, which means it interacts with the hydrocarbon chains in the membrane. Having nonpolar alpha helices allows this protein to be bound within the membrane. The purpose of being an integral protein is so that it allows the protein to generate a proton gradient by transporting protons from inside the cell to the outside of the cell. The purpose of the gradient is to allow the formation of ATP.
Each type of membrane has its specific types of lipids and more importantly proteins. The protein composition of different membranes varies even more than their lipid composition. The vertebrate retina rod cell has one part of membrane specialized for the reception of light. This specific part of membrane contains the protein, light-absorbing glycoprotein rhodopsin, which constitute 90% of the proteins of that specific part of the membrane. In a less-specialized plasma membrane like Escherichia coli membrane, there are hundreds of different proteins, including channels, transporters, and enzymes that manage metabolism, lipid synthesis, cell division and more functions.
Generally speaking, there is no specific percentage or ratio of proteins to lipid molecule. Each cell has different protein and lipid composition corresponding to its functions. Therefore, the ratio of protein to lipid molecule can range from 1:4 to 4:1.
Lipid Rafts
[edit | edit source]Structure
Lipid rafts are unique sections of membrane that have a high number of glycolipids, cholesterols, and certain proteins. The existence of lipid rafts was first proposed in 1988 by Simons and van Meer, but another set of structures known as "caveolae" were first observed much earlier. Caveolae are small depressions on the cell surface that are considered a type of membrane raft, these were named "caveolae intracellulare" (Yamada, 1955). After much debate by world renowned researchers, most in the scientific community accept that these rafts exist; however, it is unclear to most how to classify these rafts. Currently, the classification system includes three distinct groups; caveolae, glycosphingolipid enriched membranes (GEM), and polyphospho inositol rich rafts. There is also the possibility that there are inside rafts (PIP2 rich and caveolae) and outside rafts (GEM).
Most of these rafts are very highly ordered and rigid as a result of a high concentration of glycosphingolipids that characterize this portion of the membrane. The lipids included in the raft are tightly packed and extended, so they also often include other lipids with long, straight, acyl chains.
Function
Lipid rafts in the membrane have thought to carry out many important membrane functions from cholesterol transport, to endocytosis and signal transduction. Several experiments have shown that lipid rafts play an essential role in the signal transduction pathway. Although it has been proposed that the caveolae was an integral part of endocytosis, but recent data has disproved this theory. Instead caveolae are very stable regions of membranes that are not involved in endocytosis (Thompsen et al., 2002).
Rafts and the Cytoskeleton
Some actin binding proteins have been shown to bind to polyphosphoinositides and to be regulated by them by a variety of protein domains such as PH, PX and ENTH. It follows that some ABPs are suggested to link the actin cytoskeleton and PIP2-enriched rafts. One of these is gelsolin, a Ca2+, pH and polyphosphoinositide regulated actin capping and severing protein, that breaks into rafts isolated biochemically from the brain (Fanatsu et al., 2000). GEMs are also thought to link to the actin cytoskeleton through ABPs, specifically ERM proteins through EBP50, a protein that binds members of the ERM proteins through the ERM C-terminus (Brdickova et al., 2001).
References
[edit | edit source]von Heijne, G and Rees,D (2008). Current Opinion in Structural Biology. Elsevier Ltd.
Berg, Biochemistry, 6th Edition
Campbell and Reece, Biology, 7th Edition
Lehninger, Principles of Biochemistry, 4th edition
University of Edinburgh [13] All cells need to acquire the molecules and ions that they need from their surrounding extracellular fluid. There exists an exchange of molecules and ions in and out of the cell wall, as well as in and out of membrane-bounded intracellular compartments such as the nucleus, ER, and mitrochondria. Examples of substances that exchange through the membrane include glucose, Na2+, and Ca2+, ATP, mRNA, etc. The lipid bilayer of cell membranes is impermeable to large and polar molecules but permeable to water molecules and other small uncharged molecules like O2 and CO2. To solve this problem, the cell membrane contains proteins that are selective for unique, water soluble molecules. Continuous protein pathways are composed of carrier proteins, channels and pumps. The transport may be active transport by carrier proteins with an energy source, or it may be facilitated diffusion or passive transport via channels. A cell's activity depends on what biochemical reactions it may complete, and these reactions are dependent on the compounds extracted from the extracellular fluid. The array of transporters expressed in any given cell defines the cell's function and effectiveness.
Quantifying Free Energy Stored In Concentration Gradients
[edit | edit source]The process or movement of any molecule or ion moving down or up a concentration gradient requires a change in free energy. Understanding free energy is the heart of understanding how molecules are transported and/or behave in a concentration gradient. From the second law of thermodynamics molecules spontaneously move from a higher concentration to lower concentration. The availability of free energy is one of the factors that determine if a molecule will move across a membrane, the other being the permeability of the molecule in the lipid bilayer. The free energy change in the transportation of an uncharged species from one side of a membrane containing a concentration of x1, to another side of a membrane containing a concentration of x2 is given as followed:
ΔG (x) = RT ln (x2/x1) = 2.303 RT log10 (x2/x1)
R: The gas constant, 8.314 x 10−3 kJ mol−1 K−1
T: Temperature
x: Concentration
ΔG: Free Energy
At 25° (298K), ΔG is +11.4 kJ mol−1 (+2.7 kcal mol−1), indicating that this transport process requires an input of energy.
For charged species, an electrical potential is generated by an unequal distribution of ion charges across the membrane because “like” charges will be repelled. Taking the sum of the electrical terms and the concentration, electrical potential, generates the general expression.
ΔG (x) = RT ln (x1/x2) + Ζ F ΔV = 2.303RT log10 (x2/x1) + Ζ F ΔV
Ζ: Electrical charge of the transported species.
F: Faraday’s constant 96.5 kJ V−1 mol−1.
ΔV: Potential in volts across the membrane.
When ΔG is positive the transport is active, an input of energy is needed to move a molecule up a concentration gradient, contrary to ΔG being negative the transport is passive, which means that such molecules will pass through a membrane down their own gradient, simple diffusion.
Passive Transport
[edit | edit source]
Passive transport is the moving of biochemicals across membranes of cells without the use of chemical energy. The four main types in include diffusion, facilitated diffusion, filtration and osmosis.
Diffusion is the process by which molecules migrate over the cell membrane from areas of higher concentration to areas of lower concentration. When the amounts of molecules become stabilized, this state is called equilibrium. This occurs through random molecular motion.
A diffusion coefficient is a factor of proportionality that represents the amount of substance diffusion across a unit area through a unit concentration gradient in unit time. It is represented with a D and is based on Fick's law of diffusion.
There are 2 kinds of diffusion:
1) Tracer diffusion is the spontaneous mixing of molecules that occurs in absence of a concentration gradient. This diffusion takes place under equilibrium
2) Chemical diffusion occurs in the presence of a concentration gradient. It will result in a net transport of mass. The overall system will increase in entropy and it gradually moves closer to equilibrium until it is reached.
Other types of diffusion include:
- Osmosis is the diffusion of water through a semi-permeable membrane. Water will move from an area with a higher concentration of water to the other side of the membrane with a lower concentration of water. Osmosis is very important in biological systems because many membranes are semipermeable. They are impermeable to organic solutes (i.e. large molecules) but are permeable to water and small uncharged solutes. Permeability does not only depend on size, but also depends on solubility properties, charge and chemistry.Further information: [14]
- Atomic diffusion is a diffusion process that incorporates the random thermally-activated movement of atoms in a sold, which eventually results in the net transport of atoms. Examples include how a Helium atom inside a balloon can diffuse through the wall of the balloon. The balloon will eventually deflate.
- Brownian motion is the random movement of particles suspended in a fluid (could be liquid or gas). There is also a mathematical model that is used to describe such random movements.
- Collective diffusion is the diffusion of a large number of particles. This usually takes place within a solvent. This is different form the brownian motion because it is the diffusion of a single particle and the interactions between the particles may need to be considered.
- Eddy diffusion is also called "turbulent diffusion." It is any diffusion process by which substances are mixed in the atmosphere or in any fluid system due to eddy motion.
- Effusion is the process in which individual molecules will flow through a hole without collisions between molecules. It occurs if the diameter of the hole is considerably smaller than the mean free path of the molecules.
Types of Passive Membrane Transport
[edit | edit source]Ion Channels
[edit | edit source]Ion channels are proteins that forms water-filled pores through membranes, which allow specific ions to passively transport down the electrochemical gradient. For instance, the ion channels commonly found throughout the human body include Na+ channels, K+ channels, Ca2+ channels, and Cl- channels. These passive transport ion channels make use of the electrochemical potential to drive physiological processes, such as nerve impulse. The electrochemical potential is maintained by various active transport mechanisms, such as the Na+-K+ ATPase.
The selectivity of the ion channels arise from the amino acids formation from which the protein is formed. Two main criteria determine what ions pass through these channels unimpeded: diameter of the pore and the electrical properties of the amino acids. The ion channels essentially forms a hole through the lipid bilayer of the membrane. The diameter of the hole helps determine which substances are allowed to pass through the channel. Only ions of an appropriate size will be able to pass through the hole of a specific ion channel. The observation that some larger ion channels only permit a specific ion to pass while smaller ions do not is based on the energetics of dehydrating the ion as it passes. The arrangement of amino acid carbonyl groups inside channels dictate the passage: the arrangement of the carbonyl groups is so that the dehydration energy costs are lowest for certain hydrated ions. Thus a larger pore has a favorable arrangement for larger hydrated ions, whereas a smaller hydrated ion passing through would require more energy to dehydrate. In potassium channels, this dehydration region that mediates the passage of K+ ions is known as the selectivity filter.
In addition to the diameter of the pore, the electrical properties of the amino acid help determine which ions are allowed to pass through the ion channel. If the amino acids facing the inside of the pore are negatively charged, then only cations can pass through the channel. Anions will be repelled away from the negatively charged amino acids in the channel due to charge repulsion. Conversely, anions will be able to pass through channels lined with positively charged amino acids.
There are some inherited ion-channel diseases. Some include: -Chloride-channel diseases include cystic fibroses and inherited tendency to kidney stones. -Potassium-channel diseases include some inherited life-threatening defects in the heartbeat, a rare inherited tendency to epileptic seizures in the newborn and several types of inherited deafness. -Sodium-channel diseases include inherited tendency to certain types of muscle spasms and Liddle's syndrome.
Porins
[edit | edit source]
Porins are a type of membrane protein that forms a beta-barrel pore across the lipid bilayer. Unlike other protein transport channels, the pores from porins are large enough to allow passive diffusion. As a result of the large diameter, porins generally mediate the diffusion of small metabolites, such as amino acids, sugars, and ions.
The general structural properties of various porins are the same, regardless of their type. Porins are proteins composed primarily of beta-sheets, connected in the anti-parallel direction. Sixteen to eighteen strands of beta-sheets fold into a cylindrical tube, which is labeled as the beta-barrel. The amino acid sequence is roughly an alternating pattern of non-polar and polar residues, which are positioned in an appropriate way as to generate the tertiary barrel structure. This beta-barrel is located in the lipid bilayer, forming the large hole into the cell. As a result of these interactions, the amino acids facing the outside of the beta-barrel are generally nonpolar, to interact with nonpolar region of the lipid bilayer. However, the inside of the beta-barrel typically contains polar amino acids, to interact with the aqueous environment connecting the two sides of the membrane.
Porins are also found in the outer membrane of gram negative bacteria. Gram negative bacteria contain an outer membrane that helps keep out unwanted chemicals, as well as increase virulence. The porins facilitate the diffusion of small molecules and nutrients through the outer membrane and into the periplasm.
Ionophores
[edit | edit source]Ionophores are molecules that help transport ions from a hydrophilic environment into a hydrophobic environment. In other words, ionophores are ion carriers that help transport hydrophilic ions across lipid bilayer membranes. There are two broad mechanisms by which ionophores transport ions across cell membranes: carrier and channel forming.
In the carrier mechanism, the ionophore forms a complex with the ion. The ionophore wraps around the ion with its polar interior. The exterior of the ionophore-ion complex is primarily hydrophobic, thus allowing the complex to cross the hydrophobic cell membrane. The carrier ionophore basically shields the ion’s charge from the environment by solvation.
For the channel forming mechanism, the ionophore induces a hydrophilic channel through the lipid bilayer membrane. The formation of this polar pore allows ions to cross through the cell membrane. Ionophores can be used as antibiotics due to its ability to disrupt the transmembrane electrochemical gradient. This electrochemical gradient is essential in driving metabolic processes. Without the gradient, there would be no net movement of ions into and out of the cell, essentially disrupting normal cellular processes. Ionophores disrupt the electrochemical gradient by allowing ions to freely diffuse across the cell membrane, either by forming complexes or channels. This free diffusion disrupts the normal ion balance between cytoplasm and the extracellular environment, thus eliminating the electrochemical gradient.
Facilitated diffusion
[edit | edit source]Facilitated diffusion occurs in almost all cells. Facillated diffusion of ions takes place through proteins or assemblies of proteins that are embedded in the plasma membrane. These proteins are water-filled channels through which ions can pass down their concentration gradient. These channels can be either opened or closed, and others can be considered "gated".
One critical example is the metabolism of glucose. Glucose molecules cannot pass easily through cell membranes; passive diffusion alone is slow, which can be problematic for cell metabolism. One way nature has solved this problem is by facilitated diffusion (another rapid mode of glucose transport is active diffusion). Like passive diffusion, the movement of glucose is always from a region of high to low glucose concentration and independent of ion coupling. Glucose transport proteins catalyze the reaction of glucose movement across the membrane. Facilitated diffusion of glucose molecules is fast and bidirectional. The actions of glucose transport proteins are analogous to enzyme mediated catalysis. [15]
Facilitated diffusion is a form of passive transport that uses no energy for the transport of molecules and substances through the membranes of cells. It is largely aided by proteins that are an integral part of the membrane. Facilitated diffusion is the spontaneous passage of substances or molecules across a membrane passing through specific transmembrane transport proteins. The hydrophilic substances can avoid contact with the lipid by layer core of the membrane by passing through transport proteins. Some of these proteins function by having a hydrophilic channel that lets through polar molecules. Aquaporins are an example of channel proteins that facilitates the transport of water molecules through the membranes. Other proteins termed carrier proteins grab the molecules and change shapes so as to avoid contact of the molecules with the hydrophobic core and goes through the membrane. Most of the proteins are very specific that is they only transport certain types of substances.
Gated Ion Channels
[edit | edit source]Ligand-gated
These ion channels open or close in response to when it binds to a signal molecule or "ligand." The ligand is not the substance that is transported when the channel opens. Some ions are gated by extracellular ligands and others are by intracellular ligands.
Another type of protein channels are ion channels that lets through ions. Many of them are stimulated by electrical or chemical signals and are called gated channels. The stimulants either open the channel or clothe them. Stimulation of a nerve cell by certain neurotransmitter molecules, for example, opens gated channels that allow sodium ions into the cell. External ligands bind to a site on the extracellular side of the channel. Examples include:
- Acetylcholine (Ach) which is the binding of the neurotransmitter acetylcholine at certain synapses. It opens channels that admit sodium ions and initiate a nerve impulse or muscle contraction.
- Gamma amino butyric acid (GABA)- binding of GABA at certain synapses allows for the central nervous system to admit chlorine ions into the cell, which inhibits the creation of a nerve impulse.
- Calcium channels that permit calcium ions to flow into the membrane. These Ca2+ ions then serve as second messenger internal ligands to initiate various processes, such as activating calmodulin.
External ligands may be regulated by enzymes. In neurosynapses, acetylcholinesterase degrades ACh into recyclable components; GABA is degraded in a similar fashion. Ca2+ is removed by active transport via an Na+/Ca2+ pump. These various ligand removal processes help regulate the overall concentrations and restore membrane asymmetry.
Internal ligands bind to a site on the intracellular side of the channel exposed to the cytosol. Examples include:
- cyclic AMP (cAMP) ad cyclic GMP (cGMP) which are both called "second messengers". They are channels that are involved in the initiation of impulses in neurons responding to odors and light
The channel that allows chlorine and bicarbonate ions in and out of the cell is regulated by ATP. This channel is defective in patients with cystic fibrosis
Mechanically-gated ion channels
- Sound waves bending the cilia-like projections on the hair cells of the inner ear. They open up, which leads to the creation of nerve impulses that the brain interprets as sound.
- Mechanical deformation of the cells of stretch receptors open up ion channels leading the to creation of nerve impulses.
Voltage-gated channels
These neurons and muscle cells are "excitable cells". Some channels open or close as a response to changes in the charge (measured in volts) across the plasma membrane. An example is an impulse that passes down a neuron and the reduction in the voltage opens sodium channels in the adjacent portion of the membrane.
Carrier Proteins
[edit | edit source]Carrier proteins are membrane-bound transport proteins that bind with specific substrates and changes conformation to carry the substrate across the membrane. Facilitated diffusion occurs by use of carrier proteins. There are several types of carrier proteins: uniport carriers, symport carriers, and antiport carriers.
Uniport carriers are carrier proteins that move only one kind of substance across the cell membrane. They work by binding to one molecule at a time and transporting that molecule down its electrochemical gradient. Uniport carriers can be regulated by various mechanisms: voltage, physical, and ligand binding. By the voltage regulation, the uniport carrier opens or closes at a critical difference between transmembrane voltage. For the physical regulation, physical pressure will cause the carrier to open or close. Lastly, for ligand binding regulation, ligands bind to the uniport carrier on either the intracellular or extracellular side to induce opening or closing of the carrier.
Other carrier proteins can carry more than one molecule across the cell membrane at a time. Symport carriers are carrier proteins that transport two or more different molecules simultaneously across the cell membrane in the same direction. In contrast, antiport carriers are carrier proteins that transport two or more different molecules simultaneously across the cell membrane, of which at least one of the molecules are transported in the opposite direction compared to the others. Generally, these carriers transport at least one molecule down its electrochemical gradient and the other molecules would go against its gradient. The molecules going down the electrochemical gradient will provide the driving potential to push the other molecules against its electrochemical gradient. These types of proteins are classified as secondary transporters, or cotransporters.

Facilitated diffusion by carrier protein transport is ultimately based on the embedded membrane proteins, in contrast with passive diffusion which occurs everywhere in the membrane. In particular, uniport transport and passive diffusion, both of which describe ion movement down concentration gradients across membranes, differ by their maximum rates: uniport transport is limited to and located at the carrier proteins in the membrane.
Active Transport
[edit | edit source]Active transport in a cell occurs when energy is used to transport molecules across the cell membrane. While passive transport takes advantage of favorable concentration gradients to facilitate ion diffusion through membranes (i.e. moving a protein from high to low concentration), active transport requires energy input because the molecule in question is being moved against the concentration gradient (i.e. moving a protein from low to high concentration). Because this type of movement is "uphill", meaning that it is thermodynamically unfavorable, energy is needed to compensate for the thermodynamic loss. This ensures that the transport in question is completed successfully and that the cell can obtain whatever nutrients it requires, even if this means moving proteins and ions into areas where their concentration is already relatively high. The ATP used in active transportation may be used either directly or indirectly. For direct active transport, some transporters will bind ATP directly and use the energy of its hydrolysis to drive active transport and establishes a concentration gradient. Indirect Active transport will use the energy already stored in the gradient of a directly-pumped ion.
Membrane Pumps
[edit | edit source]The cell makes use of membrane pumps to help in accomplishing active transport. Pumps can convert free energy into different forms, depending on which form is required by the cell at a given time. This property makes membrane pumps a convenient choice for mediating active transport since they can provide the energy needed to initiate the transport. The two main types of pumps employed by the cell are P-type ATPases and ATP-binding cassette transporters (ABCs). Both of these pumps are powered by ATP, one of the more common forms of cellular energy. One method by which these pumps can perform active transport is by binding to ATP. This binding, followed by hydrolysis, induces a conformational change in the pump that allows bound ions to be transported across the cell membrane. These pumps can also use active transport to establish favorable concentration gradients for separate transport processes. For example, one pump can create a given concentration gradient by performing active transport on a certain ion, and then another pump can exploit this new concentration gradient by facilitating ion diffusion down the concentration gradient. Thus the cell can couple active transport with passive transport (much like it couples endergonic reactions with exergonic reactions) in order to efficiently use the results of one process to drive another process to completion.
Na+-K+ pump
[edit | edit source]
The Na+-K+ pump, also known as the Na+-K+ ATPase, is an enzyme used by the cell to control the ion gradients in its intracellular media. As the name of the enzyme suggests, the ions it deals with are those of potassium and sodium, which are two of the more common ions present in living systems. In this case, the Na+-K+ pump hydrolyzes ATP in order to supply the energy needed to actively transport Na+ out of the cell while bringing K+ into the cell. It is for this reason that most animal cells tend to have a significantly higher concentration of potassium ions than sodium ions; the cell requires this concentration in order to be able to facilitate various cellular processes. The Na+-K+ pump helps it maintain this concentration by generating the necessary ion gradient.http://www.youtube.com/watch?v=bGJIvEb6x6w&feature=related
Two other such enzyme "pumps" that are homologs of the Na+-K+ pump are the Ca2+ ATPase and the H+-K+ ATPase. Ca2+ ATPase is responsible for transporting calcium ions (Ca2+) from the cytoplasm of normal tissue cells into the sarcoplasmic reticulum of muscle cells. Calcium ions are needed by the muscle cells in order to function at peak efficiency; thus this pump ensures that they receive enough calcium ions to fulfill this requirement. H+-K+ ATPase pumps large quantities of protons (H+) into the stomach's gastric juices in order to keep the pH less than 1.0. Such a low pH is required because the stomach is responsible for digesting all the foods and fluids that enter it; thus its gastric juices need to be acidic enough to dissolve and break down anything that needs digesting.
The three aforementioned pumps are all part of a family known as the P-type ATPases, so called because they all form a phosphorylated intermediate during their reactions with ATP. Hundreds upon hundreds of known homologs of these pumps exist within the P-type ATPases, and each plays a definite role in maintaining the functions of the cell.
Multidrug Resistant (MDR) Pumps
[edit | edit source]MDR Pumps are activated by ATP and are found in microorganisms such as bacteria and cancer cells. The multidrug-resistance (MDR) pumps consist of large proteins that weave through the cell-surface membranes. They work to effectively monitor and prevent unwanted chemicals from entering the cell. Therefore, the microbes have ability to self-defend with MDR pumps. MDR pumps also prevent antibiotics from entering bacteria and chemotherapeutic agents from entering cancer cells. In addition, they are used to spit out the ones that might endanger the bacteria. Administering antibiotics with MDR inhibitors may be a way to circumvent the MDR pumps in bacteria. This explains the uncanny ability of bacteria to defend themselves against antibiotics. MDR pumps can be found in humans, where they have variety of roles that help drugs get to places they need to go. They are also important in membranes in the brain, digestive tract, liver, and kidneys because they move hormones into and out of cells through the membrane (Medicines by Design 15).
For instance, AcrB is a effective MDR in Gram-negative bacteria. It functions through its three asymmetric protomers, each of which exists as a particular conformation that corresponds to its function in the pump. The three stages are Access, Binding, and Extrusion. In the Access stage, substrates enter the protomer's vestibule, with the binding pocket still left intact. In the Binding stage, the substrate remains in the vestibule, but the binding pocket expands to better hold the substrate. Lastly, in the Extrusion stage, the substrate exits via the removal of the central helix. AcrB serves to export a multitude of drugs out of the membrane, some of which include antiseptics, toxic compounds, and antibiotics.
Vesicles
[edit | edit source]Vesicles are simply a bubble of fluid within a cell. Specifically, vesicles are a membrane bound sac that aid in transportation and storage of cellular waste or cellular products, metabolism, and buoyancy control. The membrane surrounding the vesicles shares many characteristics with the plasma membrane of the cell. As a result, the vesicle can fuse with the membrane in order to dispose of its contents.
Transport Vesicles
These are just a specific type of vesicle with a function of transport. They move molecules, products etc. between the Rough Endoplasmic Reticulum and the Golgi apparatus. Two types of proteins are synthesized on the ribosomes present on the Rough ER. From here the transport vesicles take the new proteins to the Golgi apparatus. At the Golgi apparatus the proteins age and mature in order to be transported to their final destination. The proteins always travel around the inside of the cell with transport vesicles.
Lipid Vesicles
Also known as liposomes, these vesicles are aqueous compartments that are surrounded by a lipid bilayer. These vesicles are used to monitor membrane permeability and to deliver chemicals to the cell. Lipid vesicles aid in determining the level of impermeability of a membrane to ions and polar molecules. Ions and polar molecules are trapped in the aqueous compartment of the liposome. The rate of the flow from the inner compartment of the vesicle to the outside solution during membrane transport determines the membrane's impermeability to the ions or polar molecules contained in the inner aqueous compartment. Liposomes are formed by sonicating phospholipids in the presence of an ion, polar molecules or a water soluble substance.
Coated Vesicles:
These are vesicles that form when emerging buds are detached from the cell membrane. Depending on the formation of the vesicle, the formation of the outer coat may form the vesicle, where in other cases, other enzymes are necessary to form the vesicle. It is an intracellular structure. The outer surface of the vesicle is covered in a lattice or cage like coating of the protein Clathrin.
Clathrin is a protein that forms a lattice shaped covering on the cytosolic side of the cellular membrane. These cytosolic sides are referred to as coated pits, the first stage of forming coated vesicles. It is made through a process of making subunits called triskelioins. The Triskelioins are a three pronged molecule with three N-terminus regions. They are known as heavy chains, about 192kDa in weight and are bound to light chains, about 30kDa in weight. The invagination of the pit is during the first states of endocytosis and results in a clathrin coated vesicle. Clathrin can self assemble spontaneously and it plays a major role in deforming the budding vesicle. While clathrin is a major player in the deformation, there are other accessory proteins that aid in the assembly and dissembly of the coats.
COP I is another protein that coats vesicles during protein transportation. However, this vesicle coat transports from the Golgi apparatus to the Rough Endoplasm Reticulum. Since this is in the reverse of direction of normal transportation, it is known as a retrograde transport. The protein coat is made of large protein sub-complexes that possess 7 unique subunits.
The protein is known as an ADP Ribosylation Factor dependent adaptor protein. This basically means that they are regulators of vesicle biogenesis. They also aid in the selection of the proteins that are used for the carriers.
COP II is very similar to COP I in the fact that it transports proteins, however COP II transports from the Rough ER to the Golgi apparatus and is known anterograde transport. COP II has a coat that is made of 4 unique protein subunits. COP II has three different binding sites that it can complex with other proteins.
Both COP I and COP II are very active in membrane trafficking. They perform the necessary tasks of selecting the correct cargo for the proteins and they change the shape of the phospholipid bilayer into the correct buds and vesicles.
The anterograde and retrograde transport off set each other to help keep the flow of the membranes and secretions in balance. There is constant recycling going on to maintain the equal amounts of resident proteins in the different pathways.
Exciting the Membrane
[edit | edit source]Semipermeable membranes are often sensitive to charge and voltage across the lipid bilayer. Remember, membranes are charged entities so exacting a charge across a membrane is likely to cause a reaction such as an action potential. An action potential is generally created at the axon hillock once the membrane is depolarized due to an increase or decrease of voltage. When the membrane potential goes up, integral proteins in the membrane called voltage gated ion channels start to open to allow ions into the cell. While some ion channels allow ions into the cell, other channels let ions out of the cell. This is most exemplified by the sodium potassium pump. So since sodium ions are flowing into the cell and potassium is flowing out of the cell, depolarization occurs where the usual membrane potential, called the resting potential is disturbed. Note that action potentials are all or nothing, meaning the occurrence of action potentials does not depend on the magnitude of stimulus. A single action potential can trigger several along the membrane thus propagating the signal.

Only certain types of cells have the activity of action potentials. The most important one is the neuron. A neuron is a cell in the nervous system that creates messages and sends signals through electronic excitability through action potentials. The axon hillock is the part of the neuron that connects a cell to the axon. This area has many voltage activated ion channels and is considered to be the key ingredient to creating the depolarization that exacts an action potential.

Neurotransmitters are chemicals that amplify or control the signal between a neuron and another cell. When neurotransmitters are released into the synaptic cleft, they bind to receptors and cause an action potential which can then trigger other neurons to do the same creating a surge of energy across a group of neurons that carry out a specific process. This overall process is called neurotransmission. Action potentials can be transferred from one cell to an adjacent cell because of direct connection of these cells through gap junctions. Channels and other membrane proteins make sure that action potentials only travel one direction. Some cells need no stimulus to fire action potentials while others need an external stimulus. For example, the pacemaker cells in the heart maintain a rhythm that can be changed and altered with external stimuli such as a shock to the body. While sensory neurons like the ones in your eyes that sense light are excited by external source which then provokes action potentials. This is the same for cells in the ear that are sensitive to sound.
Several chemicals block neurotransmitter binding by competitively binding to the receptors. Substances such as curare or atropine, dubbed antagonists, compete with neurotransmitters, specifically acetylcholine (ACh), binding to and preventing the ligand-gated channels from opening and allowing ions to flow, thus preventing the generation of action potentials. In a natural environment, this competitive inhibition leads to paralysis and death. In a controlled environment, these antagonists can be used to prevent muscle spasms or convulsions during surgery.
References
[edit | edit source]Alberts, B., Bray, D., Lewis, J., Raff, M. and Roberts, K. 1989. The Cell, 2nd Ed.
Berg, J.M, Tymoczko, J.L. and Stryer, L. Biochemistry. 6th ed. New York: W.H. Freeman and Company, 2007. Print.
Carruthers, Anthony. 1990. “Facilitated diffusion of glucose.” Physiological Review, vol 70-4: 1135-76. PubMedhost(accessed November 16, 2009).
Silverthorn, Human Physiology 5th Edition
Purves et al., Neuroscience, 4th Edition
http://www.biologie.uni-hamburg.de/lehre/bza/kanal/porin/eporin.htm#
http://www.answers.com/topic/ionophore
http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=mcb&part=A4038
Lodish, Harvey, et al. 2003. Molecular Cell Biology 5th Edition. pages 708-710. W. H. Freeman.
http://en.wikipedia.org/wiki/ATP-binding_cassette_transporter#Mechanism_of_Transport
Introduction
[edit | edit source]The fusion of two cells results in the coming together of the membranes of the two cells at one location which allows for the exchange of material. It is one of the most common ways for materials to enter or exit a cell and the process eventually leads to one continuous membrane surrounding the contents of the two cells. The membrane of the cells are made up of a double layer of phospholipids which interact and creates a connection between two cells. The fusion of the two cell membranes can be seen through the technique of x-ray diffraction. The technique of x-Ray diffraction confirms the theory that the cells form an hourglass shaped structure called a "stalk" during fusion. This stalk eventually grows to connect the two membranes and the two membranes eventually merge into one large one. The fusion of two membranes is catalyzed by proteins. These proteins act by recognizing other cells that have the potential to be fused and can also initiate the process by pulling the membranes close together to eliminate lipid and water interactions.
For membrane fusion to occur, the process must be thermodynamically favorable, that is, the free energy of the complex before remodeling must be lower than that of the complex afterwards. It is said that the process must be energetically "downhill" in order for the process to be feasible. The intermediate structures must also be low enough to overcome thermal fluctuations in a reasonable amount of time. The driving force of the reaction is the relaxation of elastic energy.
The amount of energy required for membrane fusion to occur is roughly 40kBT which is about the same amount of energy to hydrolyze a few ATP molecules.
Exocytotic Membrane Fusion
[edit | edit source]This is the process in which a cell's internal organelles fuse with the cytoplasmic side of the cellular membrane. It is an important process for the release of hormones and neurotransmitters as well as other biological compounds. This fusion process typically occurs in three steps. Step 1 consists of the internal cellular component that is to be released migrating towards the exterior of the cellular membrane. Step 2 is the merging of the membranes, and step 3 consists of the merging of the interior components such as molecules within a vesicle. The overall process is effected by different factors. An important factor that determines the ability of exocytotic membrane fusion is pH. Another factor is the hydrolysis of ATP as the fusion itself requires energy. Other factors include: transmembrane electrical potential, Osmotic forces, Proteinases, and Calmodulin.
Membrane Fusion Mechanics
[edit | edit source]Membrane fusion begins with the local merger of two separate cellular monolayer membranes, while the remote monolayer membranes remain separate. This initial connection between the two cellular membranes is call a fusion stalk and represents the first stage in the process of membrane fusion between two separate cellular membranes, called hemifusion. Stalk evolution eventually lead to the fusion of the remote monolayer of the two cellular membranes. The outcome of this merger is a fusion pore that connects the volumes of the previously separated cellular membranes. The fusion pore has the ability to expand or contract depending on the biological conditions. Stalk formation in the membrane fusion requires the temporary disruption of the two cellular membranes, which is unfavorable and opposed by the hydrophobic forces working to maintain a continuous lipid assembly on the cellular membrane. One theory that solves this contradiction is the involvement of one lipid molecule in the cellular membrane building a bridge between the two separate cellular membranes. In the pre-stalk fusion intermediate, one lipid molecule splays its two hydrocarbon chains into the opposing membrane and begins building a developing lipid bridge between the two separate cellular membranes. This chain-splay mechanism to counteract the hydrophobic conditions of the cellular membrane has been demonstrated countless times with simulations under conditions of partially dehydration of the membrane contact. This shows that the process of membrane fusion is ran by the strong lipid bridge force pushing between the two separate cellular membranes together. As a result of fusion, the overall curvature of the merged membrane can partially relax the bending and is reduced opposed to the former two cellular membranes. Physical forces driving unbent membranes favor membrane fusion due to the self-connectivity of two cellular membranes opposed to one small and strongly bent cellular membrane. The lipids of the merged cellular membrane can redistribute over the increased membrane area instead of being limited in one compacted cellular membrane.
Membrane Elastic Energy
[edit | edit source]In membrane remodeling there are three different energies that play a major role: membrane bending, membrane stretching, and tilting of the lipid hydrocarbon chain. Membrane bending energy is dependent on the curvature of the membrane surface. The free energy at any point on the membrane surface can be different due to the variations of lipids and protein compositions. Membrane fusion decreases the free energy of membrane bending. The second type of elastic energy is membrane stretching. This type of energy originates from the lateral tension from the membrane stretch that can drive the expansion of the fusion pore. The last type can be considered as the tilting of the hydrocarbon chain of the lipid molecules between the two cellular membranes during fusion.
SNARE complex
[edit | edit source]This is a complex made of proteins that is involved in membrane fusion. The proteins involved are called SNARE proteins which act to regulate vesicle fusion. SNARE stands for soluble N- ethylmaleimide-sensitive-factor attachment protein receptor.The SNARE complex consists of proteins that forms a four-helix bundle. They are often located on the plasma membrane and help to draw certain membranes together to initiate the fusion process. These proteins are found in all eukaryotic cells and help determine the compartment in which the vesicle will fuse. There are two kinds of SNARE proteins. The first one is the v-SNARE where the "v" stands for vesicle. The second one is the t-SNARE where the "t" stands for target. The v-SNARE which is part of the transport vesicle is an integral protein and will bind to a t-SNARE on the target membrane which will result in fusion of the target membrane to the transport vesicle. The SNARE complex plays a vital role in neurotransmission. For neurotransmission to occur, there needs to be fusion of the presynaptic plasma membrane with vesicles containing the neurotransmitter molecules. SNARE proteins must meet certain conditions in order to be active in cell fusion. The first condition is that the SNARE proteins must be located on two different membranes. Second, these membranes must be able to reach each other. Third, heavy and light chains must get together on the membrane and form t-SNAREs. Fourth, the SNAREs must be able to link the two membranes. Fifth, the anchors and linkers must be functional. Lastly, the membranes surfaces msut be compatible for fusion.
SNARE proteins can be assigned to three protein families. The families are the syntaxins, the VAMPs, and the SNAP-25 family. One of the distinguishing features that all SNARE proteins have is their coiled-coil domains. The SNARE complex itself is formed from a coil of syntaxin, a coil of VAMP, and two coils of SNAP-25. The overall mechanism of the membrane fusion is that a vesicle will land on on a membrane with the help of Rab proteins and bring the SNARE proteins closer to each other. The SNARE core complex then brings the two membranes together and creates tension within the two membranes. When the membranes get closer and closer, hemifusion can occur then a fusion pore opening and expansion allowing for membrane fusion. The SNARE proteins basically provide the driving force as well as stabilizes the transition states of the process.
One of the many important uses of membrane fusion and the SNARE complex is within the bodies nervous system. It is important for chemical synaptic transmission where neurotransmitters bound by pre-synaptic vesicles are eventually released be a calcium dependent mechanism into the synaptic cleft after membrane fusion occurs. Before fusion is able to occur, the vesicle is transported to the specific target membrane. A "fusion trigger" such as Ca2+ directs fusion for completion after the vesicle has gone through priming. In order for this fusion to occur, the SNARE complex is important in overcoming repulsive ionic forces as well as getting rid of hydration between the membranes lipid bilayers. In recent years the SNARE proteins have been reclassified as R-SNAREs and Q-SNARES instead of the v and t SNAREs. R-SNAREs are argining containing while Q-SNAREs are glutamine containing.
Hydrophobic insertion (wedging) mechanism
[edit | edit source]One mechanism of membrane fusion is the Hydrophobic insertion (wedging) mechanism. Proteins are intermediates to the process of membrane fusion and certain proteins can induce membrane fusion which in turn causes elastic stress. Examples of proteins that can drive this process are lipid-modifying enzymes, flippases and rigid protein scaffolding. The core essence of this mechanism lies in the insertion of hydrophobic or amphiphatic regions into the shallow membrane causes the expansion of the polar head region which induces protein curvature. Proteins that are capable of this include epsins, small G-proteins, and N-bar domains and proteins capable of inserting small hydrophobic loops. Further research deals with finding which structurally related proteins can drive membrane remodeling.
References
[edit | edit source]http://www.bnl.gov/bnlweb/pubaf/pr/2002/bnlpr091202.htm
http://www.lsic.ucla.edu/classes/mimg/temp/old229/vanderbliek__chen.pdf
Introduction
[edit | edit source]Membrane fission is the process in which a cellular membrane that is initially incessant is split into two distinct membranes. This activity is important in normal processes such as exocytosis and cellular reproduction and is used to separate and transport molecules from organelles with a cell to the lipid bilayer and vice versa. Membrane fission is also the direct opposite of membrane fusion and has separate proteins that will facilitate its process.
Pre-Requisites
[edit | edit source]To make a separate membrane-bound subunit without changing the original bilayer, the lipid bilayer needs to be changed and distorted locally. This must be done without creating any leakages of the carrier’s insides, thus, a fission pathway with the formation of a hemifission state is required. This hemifission state involves forming a narrow neck pathway, merging the monolayers as an intermediate, and decaying the intermediate to complete the fission reaction. The entire formation requires bending, tilting, or curving of the bilayer.
The changing of position of the lipids lead to elastic stresses, storing elastic energy in the bilayer. An outside force is needed in order to stretch, compress, tilt or bend a piece of membrane. For membrane fission to happen energetically, the total free energy (external energy plus internal energy) needs to be lower after the fission process finishes. [1]
Process and Proteins Involved
[edit | edit source]Membrane fission begins when a cellular membrane that is initially continuous begins to form a membrane neck. This neck is similar to a fusion pore and decreases in width until it fully deteriorated and the resulting two membranes form. Energy to distort the membrane and form the membrane neck is provided by the GTPase Dynamin.
The formation of the membrane neck is related to the creation of protein coats (membrane scaffolding) performed by protein complexes. The formation of protein coats can eventually lead to a membrane neck which is then narrowed by the constant construction of these protein coats. Proteins that are involved in this include CHMP2 and CHMP3 which are both charged multivesiclar body proteins. The formation of the membrane neck can also be facilitated by the insertion of hydrophobic and amphipathic proteins into or next to the cell membrane. Examples of these proteins include the BAR-domain and Dynamin family proteins which are placed onto the cell membrane.
ESCRT-III can catalyze membrane fission similarly to Dynamin family proteins except it assemblies inside the membrane which causes it to tighten and eventually start a fission. This protein is not only important for cell membrane fission, but also cytokinesis suggesting the relationship between cell membrane fission and cell reproduction.[2]
Similarity and Differences from Membrane Fusion
[edit | edit source]Membrane fission shares many similarities to membrane fusion as it is the opposite process, but also has some unique features. Both will lead to the formation of a pore like structure with a membrane neck. However for fusion, this neck will expand and eventually align linearly while for fission the neck will narrow until it is fully degraded and form two membranes are more bent in nature. The reasoning for the difference in alignment is that for fusion, the end result is a longer piece of membrane while for fission the result is two shorter pieces of membrane from the starting one.
Forces that favor membrane fission will inhibit membrane fusion while the opposite will occur for forces that inhibit it. Forces that will promote membrane bending will encourage membrane fission. This is due to the increased ability for the membrane to form the membrane neck structure created in membrane fission. Forces that decrease the membrane fluidity will have the opposite effect and as a side effect, promote membrane fusion. The membrane will not be able to form the neck used for fission as readily but the neck used for fusion in which another membrane attaches to the structure and eventually straightens out.
Dynamin family proteins, which are highly involved in several membrane fission initiations as noted, are also involved in some membrane fissions initiations. An example is in the mitochondrial cell membrane in which the Dynamin family proteins restructure the membrane by starting fission and fusion processes. For this, Dnm1 and Drp1 are the main Dynamin family proteins involved in fission while Fzo1 and Mgm1 are involved in fusion.[3]
References
[edit | edit source]- ↑ Campelo, Felix; Malhotra, Vivek (2012). "Membrane Fission: The Biogenesis of Transport Carriers". Annual Review of Biochemistry. 81: 407–27. doi:10.1146/annurev-biochem-051710-094912. PMID 22463692.
- ↑ Peel, Suman; Macheboeuf, Pauline; Martinelli, Nicolas; Weissenhorn, Winfried (2011). "Divergent pathways lead to ESCRT-III-catalyzed membrane fission". Trends in Biochemical Sciences. 36 (4): 199–210. doi:10.1016/j.tibs.2010.09.004. PMID 21030261.
- ↑ Kozlov, Michael M.; McMahon, Harvey T.; Chernomordik, Leonid V. (2010). "Protein-driven membrane stresses in fusion and fission". Trends in Biochemical Sciences. 35 (12): 699–706. doi:10.1016/j.tibs.2010.06.003. PMC 3556487. PMID 20638285.
Structure
[edit | edit source]

Micelles are lipid molecules that arrange themselves in a spherical form in aqueous solutions. The formation of a micelle is a response to the amphipathic nature of fatty acids, meaning that they contain both hydrophilic regions (polar head groups) as well as hydrophobic regions (the long hydrophobic chain). Micelles contain polar head groups that usually form the outside as the surface of micelles. They face to the water because they are polar. The hydrophobic tails are inside and away from the water since they are nonpolar. Fatty acids from micelles usually have a single hydrocarbon chain as opposed to two hydrocarbon tails. This allows them to conform into a spherical shape for lesser steric hindrance within a fatty acid. Fatty acids from Glycolipids and phospholipids, on the other hand, have two hydrophobic chains that are too bulky to fit into the a spherical shape as micelles do. Thus, they preferred to form glycolipids and phospholipids as "lipid bilayers", which are discussed in the next section.
Micelles form spontaneously in water, as stated above this spontaneous arrangement is due to the amphipatic nature of the molecule. The driving force for this arrangement is the hydrophobic interactions the molecules experience. When the hydrophobic tails are not sequestered from water this results in in the water forming an organized cage around the hydrophobic tail and this entropy is unfavorable. However, when the lipids form micelles the hydrophobic tails interact with each other, and this interaction releases water from the hydrophobic tail and this increases the disorder of the system, and this increase in entropy is favorable.
Bilayer sheet
[edit | edit source]The preferred structure of lipids in aqueous solutions are usually a bilayer sheet of lipids rather than spherical micelles. This is because the two fatty acid chains are too big and bulky to fit into the interior of a micelle. Therefore, micelles usually have one hydrocarbon chain instead of two. Lipid bilayers" form rapidly and spontaneously in an aqueous media and are stabilized by hydrophobic interactions, Van der Waals attractive forces, and electrostatic interactions. The function of the lipid bilayer is to form a barrier between the two sides of the membrane. Due to the fact that the lipid bilayer consists of hydrophobic fatty acid chains, ions and most polar molecules have trouble passing through the bilayer. The one exception to this rule is water because water has a high concentration, small size, and a lack of a complete charge. In order for a molecule to pass through the lipid bilayer it must move from an aqueous environment to a hydrophobic environment and then back into an aqueous environment.Therefore the permeability of small molecules is related to the solubility of said molecule in a nonpolar solvent versus the solubility of the molecule in water.
Micelles can also have a structure that is inside out of its normal structure. Instead of having the hydrocarbon chains inside, they can face outside and while the polar heads are arranged inside the sphere. This happens in a "water in oil" situation because there is so much oil surrounding the drop of water that the hydrocarbon chains face outside instead of inside.
Size
Sizes of micelles range from 2 nm (20 A) to 20 nm (200 A), depending on composition and concentration. The size of a micelle is more limited than that of a lipid bilayer. A lipid bilayer can span up to 107 A or 106 nm.
The lipid bilayer is not a rigid structures, rather they are quite fluid. The individual lipid molecules are able to move or diffuse laterally across the membrane quite easily, this process is called lateral diffusion. However, lipids have much more trouble flipping from one side of the membrane to the other, this process is called traverse diffusion or flip, because this would involve the polar head traveling through the hydrophobic core, and this interaction between polar and hydrophobic regions is unfavorable. So the lipid can move around laterally at a rate of about 2 micrometers per second, while it takes a much longer amount of time to flip flop.
the fluidity of a lipid bilayer also depends on both the temperature and the hydrocarbon chain. As the temperature is increased the fluidity of the lipid bilayer increases as well. Also the more cis double bonds the hydrocarbon tail has the more fluid the structure becomes. This is because when the hydrocarbon tail has cis double bonds it can no longer pack as well as the saturated hydrocarbon tail, so it becomes more fluid. Also the longer the hydrocarbon tail, the higher the transition temperature, which is the temperature at which the bilayer goes from rigid to fluid, this is because longer hydrocarbon tails can interact more strongly than shorter chains.
Formation
[edit | edit source]Micelles form when the polar head and the non polar tails arrange in a special way. They are usually driven to arrange either with the polar heads out (oil in water) or with the polar head in (water in oil). Micelles only form when the concentration of surfactant is greater than the critical micelle concentration (CMC). The surfactant is any surface active material that can part the surface upon entering. The CMC is the concentration above surfactant when micelles will form spontaneously. The higher the concentration, the more micelles there are. Micelle formation also depend on the Krafft temperature. This temperature is when surfactants will form micelles. If the temperature is below the Krafft temperature, then there is no spontaneous formation of micelles. As the temperature increases, the surfactant will turn into a soluble form and be able to form micelles from a crystalline state. The hydrophobic effect is also a driving force that needs to be taken into account. This effect is characterized by the fact that like to form intermolecular aggregates in aqueous substances and in intramolecular molecules. Micelle formation can be summed up by thermodynamics, driven by entropy and enthalpy.
Function and Usage
[edit | edit source]Micelles usually form in soap molecules. Soap often form as micelles because they contain only one hydrocarbon chain instead of two. Therefore they make up the soap property. Micelles act as emulsifiers that allows a compound that is usually insoluble in water to dissolve. Detergents and soap work by inserting the long hydrophobic tails from soap into the insoluble dirt (such as oil) while the hydrophilic head face outside and surround the nonpolar dirt. Then, this micelle can be washed away since the outside of the micelle is soluble with the solvent, which is usually polar. This is the reason why soap helps clean oily and waxy substances off from dishes since water alone cannot pull the oil off.
Micelles are also at work in the human body. Micelles help the body absorb lipid and fat soluble vitamins. They help the small intestine to absorb essential lipids and vitamins from the liver and gall bladder. They also carry complex lipids such as lecithin and lipid soluble vitamins (A, D, E and K) to the small intestine. Without micelles, these vitamins will not be able to be absorbed into the body which will lead to serious complications. Micelles also help clean the skin. Many facial washes use micelles to perform this task. They clean the skin by removing oil and other substances without the need of being washed afterward.
Also, studying membrane proteins often utilize detergents because micelles can isolate, solubilize, and manipulate them [1]
Vesicles
[edit | edit source]

Micelles show up as vesicles in biology. Unlike a micelle, however, vesicles contain a lipid bilayer, which is composed of two layers of phospholipids, arranged end to end with the hydrophobic layered buried between the two layers. A vesicle is a intracellular membrane bound sac that transports and stores substances within the cell. These vesicles store, transport, and digest waste and products from the cell. They can fuse with the plasma membrane to release things from the cell or come into the cell and put things in. Vesicles are important since they play a role in metabolism, transport, enzyme storage, and are chemical reaction chambers.

The picture above shows how liposomes are formed. The vesicles trap the glycine after sonication. Sonication disperses the phospholipids into equal size vesicles of about 500 A or 50 nm diameter sizes. The phospholipids form vesicles around the many molecules of glycine floating around. This is driven by the hydrophobic forces. After gel filtration, the vesicles are then separated from the rest of the glycine floating around. The function of this can be transport or storage of glycine to the appropriate targets. An enlarged view shows the single strand micelles around the hydrophobic glycine (Note that vesicles are, by definition, surrounded by a lipid bilayer so the image showing a monolayer of fatty acids or micelle surrounding the glycine is incorrect! Liposomes are vesicles, not micelles). The tails are inside with the glycine because they are hydrophobic while the heads face the outside which is surrounded by water.
Reference
[edit | edit source]Biochemistry, Berg
- ↑ The Journal of Biological Chemistry, Detergents as Tools in Membrane Biochemistry, November 21, 2012
Lipid bilayer is a universal component of all cell membranes. The structure is called a "lipid bilayer" because it composed of two layers of fatty acids organized in two sheets. The lipid bilayer is typically about five nanometers to ten nanometers thick and surrounds all cells providing the cell membrane structure. With the hydrophobic tails of each individual sheet interacting with one another, a hydrophobic interior is formed and this acts as a permeability barrier. The hydrophilic head groups interact with the aqueous medium on both sides of the bilayer. The two opposing sheets are also known as leaflets.
The lipid bilayer has unique properties. They are formed in sheet-like structures that contain both a hydrophilic and a hydrophobic moiety. The membrane is composed of lipids and proteins and sometimes even carbohydrates. There are two different membrane proteins in the lipid bilayer. Integral membrane proteins traverse the lipid bilayer and are adjacent to both the extracellular fluid and the cytoplasm of the cell. Peripheral membrane proteins only bind to the surface of the integral proteins and are only on either side of the membrane, outer or inner. Specific proteins on the surface of the membrane mediate different functions. For example, the sodium-potassium pump plays a major role in balancing the concentration gradient between the extracellular fluid and inside the cell. Phospholipid bilayer also has electrical properties which, as discussed with the sodium-potassium pump, allows the transfer of ions through and out of the lipid bilayer.
Structure
[edit | edit source]
The structure of the lipid bilayer explains its function as a barrier. Lipids are fats, like oil, that are insoluble in water because of its long hydrophobic tails. The hydrophobic interactions among several phospholipids and glycolipids, a certain structure called lipid bilayer or bimolecular sheet is favored. Phospholipids and glycolipids have both hydrophilic and hydrophobic moieties (amphiphilic or amphipathic). Thus, when several phospholipids or glycolipids come together in an aqueous solution, the hydrophobic tails interact with each other to form a hydrophobic center, while the hydrophilic heads interact with each other by forming a hydrophilic coating on each side of the bilayer point radically towards the polar solvent.
This lipid bilayer formation is spontaneous since the hydrophobic interactions are energetically favorable to the structure. The lipid bilayer is a noncovalent assembly. The proteins and lipid molecules are held together by noncovalent interactions such as Van der Waals forces (which holds the hydrophobic tails together) and hydrogen bonding (which binds the hydrophilic heads with water), which help to stabilize the lipid bilayer structure.
Proteins are embedded in the biological lipid bilayer membrane. The mass ratio of the lipid molecules and the proteins that are embedded in them ranges from 1:4 to 4:1. Two types of proteins exist in the lipid bilayer: integral and peripheral membrane proteins. Integral membrane proteins traverse the lipid bilayer. That is, they interact extensively with the hydrophobic region (hydrocarbon region) of the lipid bilayer. Integral membrane proteins interact by nonpolar interactions. Peripheral membrane proteins are usually attached to surfaces of integral proteins; therefore, they are on both faces of lipid bilayer. Peripheral membrane proteins interact with the hydrophilic polar head groups of the lipid molecule. Peripheral proteins bind through electrostatic and hydrogen bonds with the head group of the lipid. They usually bind to integral proteins on the cytoplasmic or extracellular side. However, they can also be covalently attached to the bilayer by a hydrophobic chain.
Lipid bilayer membranes are asymmetric, which means the outside face of membrane is always different from the inner face of the membrane.
Bilayer Dimension
[edit | edit source]The diameter of the spherical lipid bilayers starts from 250Å and goes up. The width or thickness of the bilayers is determined by the length of non polar tails. For phospholipids and sphingolipids it has ranges between 16 and 24 carbon atoms. Total width including the group heads ranges from 45Å to 50Å.
Phospholipid Translocators
[edit | edit source]Because phospholipids are synthesized from the cytosol side of the membrane, there needs to be a way to move the phospholipid towards the lumen side of the lipid bilayer. Scramblase, a translocator, is responsible for creating symmetry between the two leaflets of the lipid bilayer. As each individual phospholipid is synthesized, they are equilibrated between the layers, known as "FLIP FLOP." With the function of scramblase, it is concluded that the different types of phospholipids are evenly distributed between the two layers, seen in ER membrane.
However, in the plasma membrane, a different movement occurs. Part of the P-type pumps, flippase translocator use the energy coupled with the ATP hydrolysis to flip the phospholipids towards the cytosol side of the membrane. Therefore the bilayer in plasma membrane is maintained to be asymmetric by the flippase. Plasma membrane also does contain scramblase, but as opposed to ER scramblase, the scramblase in plasma membrane is activated only at certain times especially during apoptosis and activated platelets, where its purpose is to promote asymmetry.
Properties
[edit | edit source]Because of these interactions, lipid bilayer inherits unique properties. Lipid bilayers have "extensive" properties - they can enclose and form compartments. Lastly, they can also recover quickly if there is a hole in the lipid bilayer due to energetic reasons. However, phospholipids and glycolipids do not form micelles like fatty acids do because phospholipids and glycolipids have two hydrocarbon chains and they are too bulky to orient themselves into a sphere like a micelle. Additional properties of the lipid bilayer membrane include that they are: sheet-like, formed by lipids and proteins (sometimes carbohydrates), are amphiphatic, possess some noncovalent parts, are asymmetric, fluid, and are electrically polarized. Using fluid mosaic models, it can be seen that the bilayer undergoes rapid lateral diffusion, but flip-flop or transverse diffusion proceeds very slowly. There is also a hydrophobic transmembrane alpha helix that passes through the membrane, with the amine component on the extracellular side and the carboxy group on the cytoplasmic side.
Membrane Properties:
1. Form sheet-like structures
2. Formed by lipids and proteins (sometimes carbohydrates)
3. Lipid membranes are amphipatic (have properties that are both polar and nonpolar)
4. Specific proteins mediates membrane functions (proteins can promote permeability of membrane)
5. Non-covalent assemblies (hydrophobic forces keeps membrane together
6. Asymmetric (typically because of proteins)
7. Fluid Structures - phospholipids are constantly moving
8. Electrically polarized - has the ability to isolate/separate charges (EX: used to produce ATP or transfer nervous signals)
[1]
Functions of the lipid bilayer
[edit | edit source]The most important property of the lipid bilayer is that it is a highly impermeable structure. Impermeable simply means that it does not allow molecules to freely pass across it. Only water and gases can easily pass through the bilayer. This property means that large molecules and small polar molecules cannot cross the bilayer, and thus the cell membrane, without the assistance of other structures. This property of the lipid bilayer balance water and other organic molecules from influx/exflux through the cell and environment.
Another important property of the lipid bilayer is its fluidity. The lipid bilayer contains lipid molecules, and, it also contains proteins. The bilayer's fluidity allows these structures mobility within the lipid bilayer. This fluidity is biologically important, influencing membrane transport.
The function of the lipid bilayer membrane is mediated by the specific protein that is embedded in it. The proteins of the lipid bilayer function as pumps, channels, energy transducers, receptors, and enzymes.

Liposomes
[edit | edit source]A liposome, or lipid vesicle, is a vesicle enclosed by a lipid bilayer. Lipsomes are the opposite of micelles in that they are hollow on the inside. Liposomes are important for studying membrane permeability and they can be used to deliver ions or molecules inside cells. It is characterized by a bilayer membrane that creates an outer aqueous compartment and an inner aqueous compartment due to the hydrophilic heads and hydrophobic tails of the lipids, as shown by the figure below.

One of the ways liposomes can deliver ions and molecules to the inside of cells is through sonication of phospholipids. Phospholipids are suspended in an aqueous medium and are then sonicated, after which the phospholipids form closed vesicles. Sonication involves applying sound energy to agitate the phospholipids so that they can create liposomes around the desired ions or molecules in the solution. After the liposomes trap the ions present in the aqueous layer, gel filtration is used to wipe out the excess ions. By measuring the rate of efflux of ions from the inner compartment to the outer aqueous solution, the permeability of the bilayer can be measured. The hydrophobic layer of the membrane isolates ions from coming in-as this would be a very thermodynamically unfavorable process. This process is shown in the figure below using glycine as an example. 
Effect of Lipid Bilayers on Local Aqueous Environment
[edit | edit source]When the majority of lipids in a bilayer are anionic, they attract cations and repel anions. This will result in a special double layer solution depleted in anions and enriched in cations surround the bilayer. This layer has the ability to change the structure and function of membrane proteins. A sodium/potassium pump driven by ATP in the bilayer pumps potassium into the cell and sodium out of the cell.
Significance of the lipid bilayer
[edit | edit source]The fact of lipid bilayer, it forms a basis for cell membrane is very significant in cellular biology. If there were no membrane, there would be no distinction between intra and extracellular compartments. If there were be no cells, there would be no ways to understand how many of the biological and chemical phenomena occur since cells are the basic building blocks of life. Employing such difference between hydrophility and hydrophobicity, albeit simple, has enormous significance and consequences in the field of biochemical studies, if not the life itself. This is also an example of how intricate and fearfully the cells are made to operate to bring life.
Proteins Carry Out Most Membrane Processes
[edit | edit source]Proteins are responsible for most of the dynamic processes carried out by membranes. In particular, proteins transport chemicals and information across a membrane.
Membranes and Protein Concentration
Membranes differ in their protein content. Myelin, a membrane that serves as an electrical insulator around certain nerve fibers, has a very low concentration of proteins (18%). Relatively pure lipids are usually sufficient in their insulating properties. In contrast, the plasma membranes, or exterior membranes, of most other cells are much more metabolically active. It contains many pumps, channels, receptors and enzymes. The protein content is usually 50%. Energy-transduction membranes, such as the internal membranes of mitochondria and chlororplasts, have the highest concentration of proteins, usually 75%. In general, membranes performing different functions contain different concentrations and repertoires of proteins.
Proteins Associate with the Lipid Bilayer in a Variety of Ways
The ease with which a protein can be dissociated from a membrane indicates how intinately it is associated with the membrane. Some membrane proteins can be solubilized by relatively mild means, such as extraction by a solution of high ionic strength. Other membrane proteins are bound much more tenaciously; the can be solubilized only by using a detergent or an organic solvent. In this manner, membrane proteins can either be classified as peripheralor integral on the basis of this difference in dissociability. Integral proteins interact extensively with the hydrocarbon chains of the membrane lipids, and they can be released only by agents that compete for these nonpolar interaction. In fact, most integral membrane proteins span the lipid bilayer. In contrast peripheral proteins are bound to membranes primarily by electrostatic and hydrogen bond interactions with the head groups of the lipids. These polar interactions can be disrupted by adding salts or by changing the pH. Many peripheral membrane proteins are bound to the surface of integral proteins on either the cytoplasmic or the extracellular side of the membrane.
Secondary Structure of Membrane Proteins Proteins can span the membrane with alpha helices. The archaeal protein bacteriorhodopsin is built almost entirely of alpha helices; seven closely packed alpha helices arranged almost perpendicularly to the plane of the cell membrane, spanning its width. Examination of the primary structure of this protein reveals that most of the amino acid residues in the alpha helices are nonpolar and only a few are charged. This distribution of nonpolar amino acids is sensible because these residues are either in contact with the hydrocarbon core of the membrane or with one another. Membrane spanning alpha helices are the most common structural motif in membrane proteins. These regions can be identified with the primary sequence alone.
References
[edit | edit source]- ↑ Viadiu, Hector. "Membrane Properties." UCSD Lecture. November 2011.
1. Berg, Biochemistry, 6th Edition
2. Alberts. molecular biology of the cell, 5th edition
3. http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B7GG5-4DNGWKT-C4&_user=4429&_coverDate=10%2F29%2F2004&_alid=1119580147&_rdoc=30&_fmt=high&_orig=search&_cdi=20141&_sort=r&_docanchor=&view=c&_ct=101&_acct=C000059602&_version=1&_urlVersion=0&_userid=4429&md5=3888f90967e5fa462d6784bb99b18f4f, Encyclopedia of Biological Chemistry, pg 576-679, 12/2/2009
4. http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Fatty_Acids
5. http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Micelles
The Fluid Mosaic Model
[edit | edit source]The fluid mosaic model was originally proposed by S. Jonathan Singer and Garth Nicolson in 1972. Their idea of this model was to show and describe the general structure of a biological membrane. Biological membrane is composed of a lipid bilayer" that is essentially a two-dimensional solution composed of lipids and proteins. The lipid bilayer functions as both a solvent for integral proteins as well as a permeability barrier.
To break down each part of the name of the model: The fluidity of the model is based on the hydrophobic components like proteins and lipids. These two components allow for the membrane to have a fluid motion since it is not solid; it is not made of only one type of macromolecule. The mosaic part of the name of the model is based on the fact that mosaics are created by using different pieces to obtain an overall picture. The mosaic model consists of not just one type of integral component but rather multiple (such as glycoproteins or phospholipids). Because of the model being made of different pieces, it creates a mosaic, hence the name.
Proving the Fluid Mosaic Model
[edit | edit source]Freeze Fracture & Electron Micrographs
[edit | edit source]One aspect of the Fluid Mosaic Model is that membrane proteins are randomly distributed throughout the plane of the membrane due to their mobility (lateral diffusion). This was verified using electron microscopy to view lipid bilayers cleaved by freeze fracture.
The general outline of the technique is this:
1. Cells are frozen in liquid nitrogen.
2. Frozen cells are fractured using a knife. The fracture occurs on lines of weakness like between the lipid bilayer of the plasma membrane.
3. Freeze Etching uses a vacuum to remove surface ice.
4. The first part of making a replica is shadowing with platinum vapor at a 45-degree angle to the surface.
5. The next part of making a replica is evaporating a very thin layer of Carbon onto the surface at a 90-degree angle.
6. The final replica is revealed by degrading the organic cell material away with an acid or base.
7. The Carbon-Platinum replica is then studied under an electron microscope, and the pattern of membrane proteins is shown by the shadowed craters and bumps.
Freeze Fracture (along with metal shadowing and imaging via electron microscopy) is a technique that can be used to visualize membrane structure and protein distribution. First, a cell is rapidly frozen in liquid nitrogen. Then it is cleaved along the fracture plane that splits the lipid bilayer. Separation along this plane exposes the proteins embedded in the membrane. After fracture, the two sections are coated/shadowed with a heavy metal like platinum. Acid is used next to dissolve the organic material, resulting in a replica of the surfaces of the sample. The replicas are then viewed with an electron microscope. The micrographs show bumps on the surface of the sample, which actually are transmembrane proteins since the surfaces of the sample's halves correspond to the inner faces of the phospholipid bilayer. This confirmed that membrane proteins are randomly dispersed throughout the phospholipid bilayer, and that there are integral transmembrane proteins that span the entire membrane.
Example of technique and the resulting micrographs
Lateral Diffusion
[edit | edit source]Lateral diffusion refers to the lateral movement of lipids and proteins found in the membrane. Membrane lipids and proteins are generally free to move laterally if they are not restricted by certain interactions. Lateral diffusion is a fairly quick and spontaneous process. In this movement, cholesterol molecules move within the domain.
Lateral diffusion can be tracked by a process called fluorescence recovery after photobleaching (FRAP). This process is available because the use of fluorescence labeling allows the tracking of the molecules. The cell surface will be labeled first with a chromophore, then analyzed under a fluorescence microscope on one section (illuminated area). On this specific site, the fluoresced molecules are destroyed by bleaching them (use of laser) and watching if they leave or enter the illuminated area. If the molecules are mobile, it has two different states, bleached or unbleached.
If the molecule is leaving the illuminated area, this means that the molecule is bleached. If the molecule is entering the illuminated area, this means that the molecules is unbleached. The unbleached molecules help to increase the fluorescence intensity.
Lateral diffusion can also be measured by a complementary strategy know as fluorescence loss in photo-bleaching (FLIP). In this technique, a small area is continuously bleached and the fluorescent proteins are bleached as they diffuse into it. Eventually, the number of fluorescent proteins will decrease and will result in all bleached proteins. From both FRAP and FLIP, we can calculate the diffusion coefficient from the bleached proteins.
Although these two techniques seem promising, it has some drawbacks. One problem is that individual movement of each protein cannot be observed because there are too many bleached/fluorescent proteins. For example, one cannot tell whether each individual protein is immobile or if it's only restricted to a small area, perhaps by cytoskeleton impediment. In order to circumvent this problem, a technique known as Single-particle tracking can be used. In this technique, an individual protein is labeled by antibodies and are colored by fluorescent dye or small gold specks. The movement of these proteins are then recorded by video microscopy. Using this technique allows one to observe the diffusion pathway of a protein periodically.
Transverse Diffusion
[edit | edit source]Transverse diffusion or flip-flop involves the movement of a lipid or protein from one membrane surface to the other. Unlike lateral diffusion, transverse diffusion is a fairly slow process due to the fact that a relatively significant amount of energy is required for flip-flopping to occur. Most large proteins do not flip-flop due to their extensive polar regions, which are unfavorable in the hydrophobic core of a membrane bilayer. This allows the asymmetry of membranes to be retained for long periods, which is an important aspect of cell regulation.
In many cells, there will be protein constituents that help with the "flip-flop" process. This can be seen through comparing the flip-flop rate of a man-made lipid bilayer" versus a natural bilayer. The flip-flop rate of the man-made lipid bilayer is so slow that it can be considered to be idle as compared to the natural bilayer, indicating the existence of something in the natural bilayer that helps with process of "flipping". The flip-flop rate of phospholipids in the biological membranes is far greater than that in the artificial lipid membranes because biological membranes have protein constituents such as flipase and phospholipid translocases that accelerate the rate at which phospholipids move from one side of the bilayer to the other one.
Control of Fluidity
[edit | edit source]The fluidity of membranes determines the extent to which molecules can be transported and signals can be transducted through the membrane. Membrane fluidity is a function of its fatty acid and cholesterol content. Fatty acid chains may be ordered and rigid or disordered and fluid which affects the fluidity of the membrane in which they are contained. Long fatty acid chains are able to form stronger intermolecular interactions which restrict fluidity. Bends and kinks in the fatty acid chains formed as a result of unsaturated cis and trans double bonds may interfere with intermolecular interactions which promotes fluidity. Membrane fluidity can therefore be controlled by varying the number of double bonds and the length of fatty acid chains. Meanwhile, the presence of bulky cholesterol molecules within the membrane restrict fluidity.
Cholesterol is the key regulator of membrane fluidity in animals. It is able to interact with and form specific complexes with phospholipids that are called lipid rafts that concentrate in specific regions of the membranes. Lipid rafts result in moderation of membrane fluidity which causes the membranes to be less fluid while also making them less vulnerable to phase transitions. Cholesterol is comprised of a steroid with an -OH hydroxy group conjugated at one end and a hydrocarbon chain at the other. The rings of the steroid and the hydrocarbon chain are able to insert themselves into the phospholipid bilayer of the membrane and participate in hydrophobic interactions while the polar hydroxy group interacts with the polar head groups of the surrounding phospholipids.
Melting Temperature
[edit | edit source]The extent of fluidity can be determined by the membrane's melting temperature. As heat is increased, the membrane makes a sharp transition from a rigid state to a more fluid state. A low melting temperature indicates the presence of fatty acids that promote fluidity while a high melting temperature indicates the presence of fatty acids and cholesterol that restrict fluidity.
The melting temperature can also be affected by the ability of the molecule to pack themselves. Characteristics such as saturation, double bonds (cis or trans), and length of the fatty acid chain will affect the melting temperature. When comparing saturation versus unsaturation, the saturated fatty acid will have a higher melting temperature because the residues will react with each other causing the fatty acid to be in a more rigid state. The specific type of double bonds also affect the melting temperature. Cis double bonds will have a lower melting point compared to trans double bonds because they cannot pack themselves into a crystal as well as trans double bonds; as a result, fats with cis double bonds more readily exit the solid phase and enter the liquid phase. Longer chains of fatty acids will have high melting point because compared to shorter chains, there are more bonds to break. The more bonds present, the higher the melting point.
Other Experiments
[edit | edit source]When the fluid mosaic model was being developed, the idea that proteins could exhibit lateral movement in a membrane was a relatively new one. One of the earliest experiments to test this was performed by L. Frye and M. Edidin. Human cells and mouse cells were fused together and fluorescent labeled antibodies were used to visualize whether integral membrane proteins of each type could move among the membranes of both types in the fused cell. Rhodamine, a red fluorescent marker was used to label the antibodies specific for human proteins and the green marker fluorescin was used to label antibodies specific for mouse cell proteins. The newly fused mouse/human cells were exposed to both antibodies, and the resulting binding pattern revealed that the fused cells were not split into half red sides and half green sides, but had a intermixed content of green and red labeled proteins/antibodies. This showed that in a short amount of time, the integral proteins in fused cells were dispersed and therefore that membrane proteins have rapid lateral mobility.
References
[edit | edit source]"Molecular Biology of THE CELL." Fifth edition- Alberts, Johnson, Lewis, Raff, Roberts, Walter
Lefers, Mark. Freeze-Fracture Technique. Northwester Biology Glossary. 17 Nov 2009
http://www.biochem.northwestern.edu/holmgren/Glossary/index.html
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Fluid_Mosaic_Model
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Lipid_Bilayer
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Membrane_Lipids
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Fatty_Acids
http://en.wikibooks.org/wiki/Structural_Biochemistry/Lipids/Cholesterol
Fluid Mosaic Model
[edit | edit source]The fluid mosaic model is used to describe the interactions of lipids and proteins in biological membranes. This model essentially proclaims the concept of lateral diffusion, stating that proteins can freely move about within a membrane and that such membranes are considered to effectively be two-dimensional. The fluid mosaic model of biological membranes are always fluctuating and adjusting. In 1972, the fluid mosaic model was introduced by S. Jonathan Singer and Garth Nicholson.[1]
Fluid mosaic model of membranes states that membrane components are free to diffuse in the plane of the membrane. Some of the membrane proteins are restricted to specific regions of the membrane by interactions with cytoskeletal proteins. Also, although many phospholipids and membrane proteins can move laterally within a leaflet, they do not flip-flop from one leaflet of the bilayer to the other. Flip-flop of the phospholipids is very rare. The inner and the outer leaflets of the membrane may be made up of different phospholipids. Membrane fluidity refers to the movement of membrane phospholipids within the plane of the membrane. A decrease in fluidity is associated with decreased transport rates. The length of the fatty acid side chains also affects fluidity. The phospholipids with long hydrocarbon chains have increased hydrophobic interactions with neighboring lipids and thus decreased membrane fluidity. [Microbiology]
Some organisms can alter membrane fluidity in response to temperature stress by changing the length and degree of saturation of fatty acids present in membrane phospholipids. Cholesterol also influence membrane fluidity. The effects of cholesterol on membrane fluidity are complicated and depend on factors such as the ratio of saturated to unsaturated fatty acids in the membrane. The cholesterol also prevents packing of saturated fatty acids, thus increasing fluidity.
Proposed by S.J. Singer and Garth L. Nicholson in 1972, the fluid mosaic model provides a reasonable structure and image of the biological membranes in general. One of the most important features of this model is the idea that the phospholipid bilayer is fluid. The phospholipid molecule are free to move laterally. Relative to the lateral movement of the phospholipid molecules, there is very little exchange between the two halves of the bilayer. This minimal exchange, or flip flop action, allows asymmetric distribution of phospholipids. This asymmetry is an important feature of membranes. Membrane surfaces exhibit asymmetry. In other words, they have different characteristics on the two sides. These structural differences support the functional differences of the inner and outer sides of the membrane. For example, one of the most important functions of the outer surface of the membrane lies in its interaction and communication with other cells. This is often achieved by sugar molecules almost exclusively found on the outer surface that acts as distinguishing markers for the cell. The interior, on the other hand, serves different functions, and therefore has a different composition. In this model, the membrane is a mosaic of proteins embedded in a fluid phospholipid bilayer. The hydrophilic portions of the phospholipid and proteins are maximally exposed to aqueous interface. This feature ensures membrane stability. The fluidity of the molecule is affected by several factors. These include the type of lipid found in the membrane and the degree of unsaturation in the fatty acid chains of membrane lipids. The presence of a cis double bond introduces a kink into the fatty acid chain, which affects the packing of the phospholipid bilayer. The kink prevents the phospholipid molecules from being packed together too tightly, and thus contributes to the membrane fluidity. It is important to understand that in this model, both the membrane lipids and the embedded proteins are free to move. They may be mobile or fluid.
Proof of Fluidity
[edit | edit source]The fluid mosaic model obviously states that the lipid bilayer that surrounds the cell is fluid, flexible and always moving. In order to prove that an iconic experiment was done by taking a cell and saturating the lipid bilayer with fluorescence. After the cell was completed saturated with a green fluorescense, the cell was bleached in a single spot in the cell membrane. This created a very white spot among the green fluorescence coated cell membrane. After a short while, the bleach spot began to diminish in color and before you knew it, the area seem to be recoated with green fluorescence. The phenomenon behind this was the aspect of the cell membrane diffusing the bleached hydrophilic heads amongst the rest of the cell membrane. This diffusion allowed for the white bleach to diminish in color by being substituted by green fluorescenced hydrophilic heads. There was found to be two types of diffusion in the cell membrane, lateral diffusion and transverse diffusion. Lateral diffusion is the switching of positions in a side by side manner without any sort of flipping. This is the fastest mode of diffusion found in the cell. the second diffusion method transverse diffusion, is the flipping of the phospholipid heads to either side of the cell membrane. This method of diffusion however is less likely and happens significantly slower then lateral diffusion.
History & Development
[edit | edit source]Singer's studies of membranes started in the 1950s when scientists noticed that many water-soluble proteins (like those in cells) were also able to dissolve in nonpolar, nonaqueous solvents and that proteins adopted different shapes in hydrophilic vs. hydrophobic environments. Many proteins are found in environments also high in lipid content, and this prompted Singer to look into the relationship between proteins and lipid membranes.
Before Singer and Nicolson's fluid mosaic model of membranes, a triple-layered membrane model was proposed, the Davson-Danielli-Roberston (DDR) model. This model proposed a triple-layered membrane, with a lipid layer between two flat protein layers. However, when studied with respect to energetics of hydrophobic/hydrophilic interactions, this model is not feasible. Due to hydrophobic and hydrophilic interactions between amino acid residues of a protein, Singer therefore proposed that membrane proteins would assume folded conformations, not remain in a flat layer like the DDR model proposed. Also in contrast to the DDR model, Singer also proposed that logically and for maximum stability membrane proteins would not be separated from the lipid bilayer but rather incorporated as part of the membrane. [2] These conclusions all came together in the fluid mosaic model, where the phospholipid bilayer is a fluid matrix and both lipids and proteins are capable of lateral and rotational movement (see membrane fluidity).
References
[edit | edit source]- ↑ Nelson, David L, Michael M. Cox. Lehninger Principles of Biochemistry Fourth Edition. New York: W.H. Freeman, and Company 2005.
- ↑ Martin, Laura. The Fluid Mosaic Model of the Cell Membrane - The Mosaic. Connexions. 15 Oct. 2007 <http://cnx.org/content/m15255/1.2/>.
Slonczewski, Joan L. Microbiology "An Evolving Science." Second Edition. There are many different techniques to study membranes. Four important techniques are:
- Electron Microscopy
- Preparation of Bilayers and Vesicles
- Scanning Calorimetry
- Fluorescence Photo Bleaching
Electron microscopy is a technique used for gaining a molecular picture of matter. The resulting picture of a compound is called an electron micrograph. Depending on the resolution of the device, micrographs can show images of particles and macromolecules on the nanometer scale. This is particularly effective in biochemistry because electron micrographs can give images of a particular protein at different angles and allow a three dimensional image of the outside of a protein to be created. This image gives an insight into the structure, and thus the function, of the protein in question.
Electron microscopy works on the basic principle of shooting a beam of electrons at a particular material (often under vacuum to avoid electron collisions with air molecule) and "reading" the result. The particular way a result is "read" gives different types of images for different materials and classifies the different types of electron microscopy. These different types are Scanning Electron Microscopy (SEM), Transmission Electron Microscopy (TEM), Scanning Transmission Electron Microscopy (STEM), Reflection Electron Microscopy (REM), and Low Voltage Electron Microscopy (LVEM).
Scanning Electron Microscopy (SEM)
[edit | edit source]
SEM is a type of electron microscopy that images a sample's surface by scanning it with a high-energy beam of electrons in a rectangular formation of parallel scanning lines that is similar to the guidance of an electron beam on a television screen or a computer monitor. The electrons interact with the atoms that make up the sample producing signals that contain information about the sample's surface topography, composition and other properties such as electrical conductivity. Scanning electron microscopes usually have a magnification range from 15x to 200,000x and a resolution of around 5 nanometers.
Just recently, in December 2012, the Italian scientist, Enzo Di Fabrizio, has obtained the first real photo of the DNA double - helix structure. In the image, one can see seven strands of DNA wrapped into a cord. Di Frabrizio used electron microscopy, which is a very useful technique to obtain a molecular picture at a nanometer scale. What he did was "developed a technique that pulls strands of DNA between two miniscule silicone pillars, then photographs them via an electron microscope"(Grenoble). This is a great example of how well electron microscopy works to obtain images of particles and macromolecules.

Transmission Electron Microscopy (TEM)
[edit | edit source]TEM uses a high voltage electron beam to create an image. An electron gun is then used to emit these electrons, which is usually fitted with a tungsten filament cathode as the electron source. The electron beam is accelerated by an anode/cathode mechanism, which is then focused by electrostatic and electromagnetic lenses, and transmitted through the specimen that is transparent to electrons and in part scatters them out of the beam. Upon leaving the specimen, the electron beam contains information about the structure of the specimen that is visually enhanced by the microscope. The image is viewed by projecting the magnified electron image onto a fluorescent viewing screen coated with a phosphor or scintillator material such as zinc sulfide.
Scanning Transmission Electron Microscopy (STEM)
[edit | edit source]STEM is a type of transmission electron microscope, where the electrons pass through the specimen, but, with scanning electron microscopy, the electron optics focus the beam into a narrow area which is scanned over the sample in a raster. This process makes these microscopes suitable for analysis techniques such as mapping by energy dispersive X-ray (EDX) spectroscopy, electron energy loss spectroscopy (EELS) and annular dark-field imaging (ADF). These signals can be obtained simultaneously, allowing direct correlation of image and quantitative data. By using this process, it is possible to form atomic resolution images where the contrast is directly related to the atomic number.
Reflection Electron Microscopy (REM)
[edit | edit source]In REM, an electron beam is incident on a surface, but instead of using the transmission (TEM) or secondary electrons (SEM), the reflected beam of elastically scattered electrons is detected. This technique is usually coupled with Reflection High Energy Electron Diffraction (RHEED) and Reflection high-energy loss spectrum (RHELS). This method improves clarity by increasing the resolution and increasing light scattering for a greater sensitivity in structural differences. This method is particularly effective in identifying molecular structure since electrons reflected can have their distance measured and calculated through geometric manipulation. However, reflective methods may not be as effective in certain environments since the index of refraction may not be constant if performed outside a vacuum.
Low Voltage Electron Microscopy (LVEM)
[edit | edit source]The low voltage electron microscope (LVEM) is a new type of microscope that is used to observe biological specimens. LVEM is divided into 2 parts: a miniature transmission electron microscope and conventional optical microscope. The miniature transmission electron microscope has maximum magnification 500 times by using emitter source of Schottky type, magnetic lens (for image formation), and electrostatic lens (for controlling magnification). The single crystal YAG fluorescent screen converts the electron image to light image. The miniature transmission electron microscope only uses 5kV source for accelerating voltage. The electron microscope is kind of small in size (around 20cm) and fits into a conventional optical microscope which has maximum magnification 400 times. The ability of having that maximum magnification is due to the aid of CCD camera (for image recording). The low accelerating voltage 5kV helps LVEM having imaging contrast twenty times higher than for 100kV. This seems to be an advantage of LVEM. However, the disadvantage of LVEM is a low transmittable thickness of the sample below 20 nm which limits only for small objects with the size of 20 nm.
References
[edit | edit source]Grenoble, Ryan. "DNA Photo Shows Double Helix For The First Time." The Huffington Post. 2012. Calcium cation is required for the process of lipid vesicle fusion to form a phosphate lipid bilayer. Scanning Calorimetry is a technique used to study phase transitions in lipids as well as proteins and other non-organic systems. The basic idea of scanning calorimetry is the use of two cells one containing a buffer of known specific heat and the other to be studied. In scanning calorimetry, heat is added to both cells while the temperature of both are being recorded. When temperature differences occur, the calorimeter understands to add heat to one of the cells so that the temperature in both cells are always equal. The scanning part comes from the constant scanning/measurement of temperature and heat, dependent on time. As the cell being studied reaches a phase transition, it will begin to need much more heat to raise the temperature of the studied cell to that of the sample cell.
Applications in Biochemistry
[edit | edit source]Scanning calorimetry can be used to study the melting temperature,Tm of proteins and lipids. By using the same principles as mentioned in the overview, it can be determined when a protein is becoming denatured. Also when equations of enthalpy and Gibbs free energy are combined with calorimetry, the relative amounts and specific heats of proteins, lipids, and other macromolecules can be determined. Scanning calorimetry has allowed scientists to elucidate structures and interacts occurring in macromolecules quantitatively, that otherwise would not be known. Such values as stability in native form can be measured by Scanning calorimetry, by using the equation change in G=-RT(lnK) where G is the gibbs free energy, R is the gas constant, T is the temperature in Kelvin, and K is the equilibrium constant.
Liquid Crystals
[edit | edit source]Scanning calorimetry can be effectively applied to study phase shifts. When a solid enters a liquid phase it is not simply a discrete transformation but rather one with a transition state and a gradual process involving kinetics atom arrangement. The state that is occupied between a solid and a liquid is called the mesomorphous state and by using scanning calorimetry we can observe energy changes in a given period of time and relate that energy change to how rapidly or slowly the atoms are dispersing and rearranging themselves.
Oxidative Stability
[edit | edit source]Using an airtight chamber scanning calorimetry can be used to determine measures of storing a certain compound or the chemical conditions necessary for a successful procedure or investigation. Oxidative tests using nitrogen can be added to a system with the compound of interest. If oxidation occurs and depending on the rate and we can readily identify if one condition is suitable or not suitable to store a sensitive compound. Therefore scanning calorimetry makes a good safety tool. By studying an exothermic event and assessing the stability of a certain compound in different amounts of heat scanning calorimetry can tell us the maximum temperature of the material and its characteristics of its micro chemistry under heat.
Polymers
[edit | edit source]Polymers are often broken down to identify different parts of their composition. Scanning calorimetry can check the composition using databases and compilations of standards. Scanning calorimetry of individual substituents can help identify parts of complex polymers that we find the future. By degrading a polymer we can also investigate the heat and exothermic character once again and then we can tell what the composition is made of. Scanning calorimetry also provides graphs that can show crystallization peaks which can tell us the percentage of crystallinity which will give us insight on polymer purities and freezing point depression. And of course this can be applied to medicine because scanning calorimetry can assess what temperature to process a drug so the drug does not crystallize and become untakeable. Also cross linking of polymers is common in polymer industries and each time polymers do so, scanning calorimetry can provide a graph curve to show the time of transition and that can be further evaluated.
Introduction
[edit | edit source]As briefly stated in a previous section on the fluid mosaic model of biological membranes, proteins and phospholipids diffuse both laterally and, to a lesser extent transversely, through the entire span of a membrane. This sort of behavior can be characterized by fluorescence microscopy. This particular technique is called FRAP or fluorescence recovery after photo bleaching. In a typical procedure, a specific portion of a cellular membrane is first tagged with a fluorescent chromophore. Next, an intense light is pulsed over a small part of the fluorescent-marked region and viewed under a microscope. As a result of the exposure to the powerful laser-light, the fluorescent molecules are bleached (destroyed). The bleached region is then monitored over time for the recovery of fluorescence (as neighboring unbleached molecules moving towards the bleached areas). This can determine the availability state of a protein - whether it is free to diffuse or is already bounded. The rate at which the recovery takes place, D, the diffusion coefficient, will reflect the overall mobility of membrane components. The equation for such observations is as follows where S is the average distance of movement per unit time (t) as they relate to the diffusion coefficient:
S = (4Dt)1/2

A typical speed for a membrane phospholipid is 2um per second. This may not sound very fast, however, if you take into account the relative length of a cell or organelles which are typically 2um or less, a phospholipid can easily traverse an entire surface in 1 second or less. This experiment also allows for comparison with other familiar materials where relative diffusion rates and viscosities are known. As it turns out, a membrane has a diffusion rate and viscosity similar to that observed in olive oil. The speeds for membrane proteins vary a great deal from nearly immobile to nearly as fast as a phospholipids. This should not be surprising being that proteins have, in general, a more irregular shape and a more uneven distribution of polar and nonpolar groups. Indeed, some of the more immobile proteins are anchored to various structures within the cell.
GFP
[edit | edit source]
GFP is a protein that exhibits a green florescent light when exposed to blue light, hence its name. Green florescent protein was the first protein extracted from A. Victoria. Other marine animals, mostly corals, also exhibit florescence. The color of the florescence’s varies based on the structure of the derivative.
GFP Uses
[edit | edit source]
GFP's can be used to study certain cell processes that could be difficult if not impossible to study. Including DNA replication and transcription, protein synthesis and transport, organelle and cytoskeleton assembly/disassembly, energy metabolism, and signal transduction pathways. Fluorescence is useful for measure cellular processes over time scales of minutes seconds or hours.. The most important aspect of GFP is it is genetically encoded by a single portable DNA sequence. This can be covalently bonded to a protein that can then be expressed in vivo.

FRET
[edit | edit source]
Florescence Resonance Energy Transfer has two major contributors. A donor chromophore and an acceptor chromophore. This allows for protein-protein interactions to be resolved spatially and temporally.The mechanism of fluorescence resonance energy transfer involves the use of a donor fluorophore. The fluorophore must be in an excited state. In the excited state the fluorophore may transfer its energy to a nearby acceptor chromophore. The main idea behind fret is that the energy transfer is based on the concept of treating an excited fluorophore as an continuously oscillating dipole. This can be compared to pair of tuning forks vibrating at the same frequency. Resonance energy transfer can yield a significant amount of structural information concerning the donor-acceptor pair.
The "phenomenon" of fluorescence resonance energy transfer is not regulated by photon emissions. FRET does not need the acceptor chromophore to be fluorescent either. The efficiency of the energy transfer varies in a direct proportion to the distance separating the acceptor and donor. FRET measurements can also be used as a ruler in molecular scale to determine distances between molecules. They must be labeled with an appropriate donor and acceptor fluorophore and must be within 10 nanometers of each other.
Credits
[edit | edit source]Lippincott-Schwartz J Annu Rev Biochem.2011 Jun 7 ; 80():327-32.
Berg, Jeremy, Tymoczko J., Stryer, L.(2012). Photobleaching Steps.Biochemistry(7th Edition). W.H. Freeman and Company. ISBN1-4292-2936-5
Waxes
[edit | edit source]Chemical Structure
[edit | edit source]Wax is a type of long chain non polar lipid which made up of various n-alkanes, ketones, primary alcohol, secondary alcohols, mono esters, beta di ketones, aldehydes, etc. Waxes will form protective coating on plants and fruits, and in animal (example: beeswax, whale spermatics, etc.). More commonly, wax is ester of alcohol and fatty acids. They differ from fats since they don’t have triglyceride ester of three fatty acids. Waxes are water resistant, so they are insoluble in water.

Properties
[edit | edit source]Due to the versatility of waxes, nature has manipulated them for their water-resistant properties, colligative properties (high melting point, relatively low viscosity at high temperatures, transparency, etc.) and coating properties.
Types of Waxes
[edit | edit source]- Beeswax – for consumption
- Chinese Wax – for polishes
- Ear Wax – used as a protective layer over the ear canal
- Lanolin – for rust prevention and cosmetics
- Shellac – used as a wood sealant
- Spermaceti – for cosmetics and leather works
- Vegetable (many different types extracted from plants) – used as a protective layer on the plant to prevent loss of water
- Mineral – used as fine polishes
- Petroleum – fuels, paints, culinary, candles
- Synthetic – modified waxes for use in the medical field
Functions and Applications
[edit | edit source]Waxes contain many functions in society. Man has manipulated and synthesized many waxes to be used for cosmetics, sealants and lubricants, insecticides, UV protection, energy reserves, food, etc.
Soaps
[edit | edit source]Soap is a mixture of sodium salts forming by adding sodium hydroxide or sodium carbonate to natural fatty acids. The density of soap will be decreased by air bubble forming. The general reaction in producing soap:
- Fat + NaOH → soap + glycerol
Making Soaps
[edit | edit source]An example of making soaps is called Saponification of fats and oils which is the most common method of soap making process. 1) Heat fats and oils 2) React with liquid Alkali

Detergents
[edit | edit source]Detergent is a material that helps in cleaning. Detergent contains one of more surfactants which are capable of reducing the surface tension of liquid such as water. Commonly, detergent consists of long chain hydrocarbon and ionic group (such as alky sulfate or derivative of ammoniac group).
Types of Detergents
[edit | edit source]Anionic
These detergents are man-made and consist of long hydrocarbon chains and a water-soluble ionic group, which is usually negatively charged. These detergents are commonly known as surfactants, or alkyl benzene sulfonates
Cationic
These detergents are also man-made, and they only differ from anionic detergents in that the water-soluble ionic group is positively charged. These detergents are primarily derivatives of ammonium and are commonly used as a germicide and in shampoo.
Neutral
These type of detergents contain the same general set up as all other detergents, except it’s overall charge is neutral. The head of the detergent is polar due to the presence of three hydroxyl groups and an ester group.
Natural
Natural detergents such as bile salts (sodiumglycoholate) are made in the liver. These detergents are derivatives of cholesterol, a type of lipid. Its main function is digestion. The bile salt is capable of emulsifying fats and oils such that enzymes may break them down further.
Function and Application
[edit | edit source]Detergents have many functions and applications including cleaning off fats and oils, softening materials, acting as aromas or abrasives, or sanitizing and disinfecting. Though detergent and soap are used interchangeably, people can see the various uses of detergents in common house products including hand soap, laundry detergent, dish and glass detergents, etc.
Fatty Wax
[edit | edit source]A wax is a simple lipid that is an ester of a long-chain alcohol and a fatty acid. The alcohol may be made up of 12-32 carbon atoms.
These waxes can be found in nature as coatings on leaves and stems of plants, and prevents the plant from losing excessive amounts of water. Carnuba wax is found on the leaves of Brazilian palm trees and is used in floor and automobile waxes. Lanolin coats lamb's wool. Beeswax is secreted by bees to make cells for honey and eggs. Spermaceti wax is found in the head cavities and blubber of the sperm whale. Many of these waxes are used in ointments, hand creams, and cosmetics, which can be seen in the ingredients lists.
Paraffin wax which is used in some candles, is not based upon the ester functional group, but instead is a mixture of high molecular weight alkanes. Ear wax, on the other hand, is a mixture of phospholipids and esters of cholesterol.
The waxes with their component alcohols and fatty acids are listed below:
Carnuba:
Alcohol- CH3(CH2)28CH2-OH, Fatty Acid- CH3(CH2)24COOH
Beeswax:
Alcohol- CH3(CH2)28CH2-OH, Fatty Acid- CH3(CH2)14COOH
Spermacetic:
Alcohol- CH3(CH2)14CH2-OH, Fatty Acid- CH3(CH2)14COOH
Ester Synthesis:
Simple esters contain an organic acid and an alcohol. The ester functional group is of significance in the biochemical group of compounds called waxes, triglycerides, and phospholipids. The simplified reaction reveals the process of breaking some bonds and forming the ester and the by product, water. It is as follows: First, the -OH bond of the acid is broken as well as the -H bond of the alcohol. Both join to make HOH, a water molecule. Secondly, the oxygen of the alcohol forms a bond to the acid at the carbon with the double bond oxygen. This forms the ester functional group. Note that the long carbon chains do not participate in the reaction, but are just part of the final molecule.
Lipstick:
Lipstick is a common form of fatty wax. It consists of a suspension of coloring agents in high molecular weight hydrocarbons, waxes, and/or fats. The color usually comes from a dye precipitated by a metal ion such as Fe (III), Ni(II), or Co(II) ions. An ingredients list on the lipstick might contain: dye (4-8%); castor oil, paraffin, or fats to dissolve dye (50%); lanolin (25%); carnauba and/or beeswax as a stiffening agent (36%); perfume (1.5%). The lipstick is made by first dispersing the dye in the castor oil. Then, the other waxes and lanolin are added as the mixture is heated and stirred. The molten waxes are then cast in a suitable form to harden.
Eye Makeup:
Eye makeup is also another form of fatty wax. Eyebrow pencils are similar to lipstick but contain lamp black (carbon soot) as a black coloring agent. A different mixture of waxes may be used to give the desired melting point. Brown pencils are made by adding iron oxide (rust) as a pigment. A water-resistant mascara has a mixture of waxes, fats, oils, and soap. Other coloring agents in addition to blacks and browns may be chromic oxide (dark green) and ultramarine (blue pigment of sodium and aluminum silicate).
Reference: Ophardt, Charles E. Elmhurst College, Virtual Chembook 2003. http://www.elmhurst.edu/~chm/vchembook/554wax.html
Soap
[edit | edit source]Soap is a form of lipid which is a mixture of sodium salts of various naturally occurring fatty acids. When air bubbles are added to a molten soap, the density of the soap decreases thus making it float on water. A softer soap results when the fatty acid salt contains potassium rather than sodium. Soap is the product of a saponification or basic hydrolysis reaction of a fat or oil. Sodium carbonate or sodium hydroxide is currently used to neutralize the fatty acid and convert it to the salt.
The basic hydrolysis reaction is as follows:
- Fat + NaOH → sodium salt of fatty acid + glycerol
This reaction occurs in two steps and the net effect is the broken ester bonds. The glycerol first turns back into an alcohol, then the fatty acid portion is turned into a salt due to the presence of the basic NaOH solution. The oxygen of the carboxyl group now has a negative charge which attracts the positive sodium ion.
Structure
[edit | edit source]Micelles are formed when a certain molecule are added with water. The molecule is fatty acids, phospholipids, or a salt of a fatty acid (soap). The molecule has a strong polar head and non-polar hydrocarbon tail. When this molecule is added to water, the non-polar tails associate each other in the center like a ball because their hydrophobic tails or “water hating” are not soluble in water. The polar heads of the molecule form a shell outside of the ball and interact with the water molecules.
Types of Soap:
The unique properties of different soaps are based on the type of fatty acid and length of the carbon chain of the molecule. Tallow or animal fats give primarily sodium stearate (18 carbons)- a very hard, insoluble soap. Fatty acids with longer chains are even more insoluble. As a matter of fact, zinc stearate is used in talcum powders because it is water repellent.
Coconut oil is a source of lauric acid (12 carbons) which can be made into sodium laurate. This soap, on the other hand, is very soluble and will lather easily even in sea water.
Fatty acids with only 10 or fewer carbons are not used in soaps because they irritate the skin and have objectionable odors.
Cleansing Action of Soap:
The cleansing action of soap is determined by its amphipathic properties- polar and non-polar structures, as well as its solubility characteristics. The long hydrocarbon chain is non-polar and hydrophobic (repelled by water); and the "salt" end of the soap molecule is ionic and hydrophilic (water soluble).
Monolayer: As soap is combined with water, the ionic-salt end of the soap molecule is attracted to the water molecules and consequently dissolves in it. The non-polar hydrocarbon end of the soap molecule is repelled by the water molecules. A drop or two of soap in water forms a monolayer on the water surface. The soap molecules "stand up" on the surface as the polar carboxyl salt end is attracted to the polar water. The non-polar hydrocarbon tails are repelled by the water, which makes them appear to stand up.
Soap vs. Oil vs. Water:
Due to their opposite polarity, water by itself cannot penetrate grease or oil. However, when grease or oil (non-polar hydrocarbons) are mixed with a soap-water solution, the soap molecules work as a "bridge" between polar water molecules and non-polar oil molecules. Soap molecules are amphipathic and thus have both properties of non-polar and polar at opposite ends of the molecule.
The oil is a pure hydrocarbon so it is non-polar. The non-polar hydrocarbon tail of the soap dissolves into the oil. That leaves the polar carboxylate ion of the soap molecules are sticking out of the oil droplets, the surface of each oil droplet is negatively charged. As a result, the oil droplets repel each other and remain suspended in solution (this is called an emulsion) to be washed away by a stream of water. The outside of the droplet is also coated with a layer of water molecules. This is also similar to a micelle which works with the same principles- the center of the micelle would contain the oil.
Effect of Hard Water:
When soap is used in "hard" water, it will be precipitated as a "bath-tub ring" by calcium or magnesium ions present in the "hard" water. The effects of "hard" water calcium or magnesium ions decrease with the addition of "builders". The most common "builder" is sodium trimetaphosphate. The phosphates react with the calcium or magnesium ions thus keeping them in solution but away from the soap molecule. The soap molecule can then do its job without interference from calcium or magnesium ions. Other "builders" include sodium carbonate, borax, and sodium silicate, which are currently in detergents.
Shampoo:
Water is the primary ingredient in shampoo, making up 60-70% of its content. The ingredients in shampoo are most often biodegradable. Detergents are the next most abundant ingredients in shampoo. Detergents are surfactants that react with the surface in order to remove oil and dirt particles from the hair follicle. The insoluble portion aligns with the hair’s oil particles while the water soluble portion aligns on the outside, creating a micelle. Common detergents include ammonium laurel sulfate, sodium laurel sulfate, and sodium laurel ether sulfate. Sodium laurel ether sulphates are a popular ingredient in shampoos because of their biodegradability and ability to give a “foaming” effect. The sulphates are made from either natural or synthetic linear C12-C15 alcohols. Thickeners are then added in order to increase viscosity. Popular thickeners include sodium chloride and methylcellulose. Shampoo strips the hair of its sebum, which is required to protect the hair shaft. Therefore, shampoos must also contain an ingredient to replace the lost sebum. Silicones, polymers, and quaternary agents coat the hair and replace the lost sebum. Because shampoos consist of mostly water and organic compounds, preservatives such as parabens are added to prevent the growth of bacteria and maintain freshness.
References
[edit | edit source]http://nzic.org.nz/ChemProcesses/detergents/11A.pdf http://www.madehow.com/Volume-3/Shampoo.html
Ophardt, Charles E. Elmhurst College, Virtual Chembook. 2003 http://www.elmhurst.edu/~chm/vchembook/554soap.html
Chemical Structures
[edit | edit source]Detergents are primarily made up of surfactants, or “surface active agents” of which reduce the surface tension of water by adsorbing at the common boundary between liquid and gas or one liquid and another. Surfactants form into aggregates known as micelles, formations of amphiphilic lipids. In micelles, the amphiphilic lipid has a tail that forms a core that encapsulates an oil droplet or dirt particle and a head that maintains contact with the surrounding water environment. To work effectively, the chemical formation of micelles is not enough to remove oil or grease; mechanical energy (scrubbing or water flow) is often required.
Surfactants can be formed from petrochemical or oleochemicals, chemical products made from raw materials such as petroleum or other hydrocarbon substances (olefin or aromatics). Alkalis and oxidizing agents are other components of detergents. Alkalis provide positively charged cations that can trigger chemical reactions. Oxidizing agents can act as a source of energy for chemical reactions and include sulfuric acid and ethylene dioxide.
Detergents can also be composed of substances that can modify pH of other components, as well as water softeners (compounds that reduce metal ion concentration) and enzymes that digest proteins or fats. In addition, detergents can comprise of ingredients that modify the presence of foam or stabilize the viscosity of a solution.
CH3(CH2)14-CO-O-(CH2-C-OH)3
Properties
[edit | edit source]•Nonionic detergents, which form electrically neutral colloidal particles in solution, contain non-ionic emulsifiers, amplifying the ability to remove oily residues. These nonionic surfactants are low-foaming and have defoaming properties that improve wetting, rinsing and particle removal while not hindering with mechanical labor. A common nonionic detergent is Liquinox used to clean glassware and does not react with hard water ions; this is due to the fact that no ionic groups exist to do so. In addition, it foams less than ionic detergents but has some polar portions to provide necessary water solubility.
•Cationic detergents often come in powder foam and have a long cationic chain that is responsible for surfactant properties. However, cationic detergents are poor detergents even though they have adequate emulsifying properties. Instead, there are often used in bacterica/germ free environments due to their antiseptic properties.
•Anionic detergents are based on sulfate or carboxylate anions and are of the most common modern synthetic detergents producing negative charged colloidal ions in solution.
•Bile salts in the stomach have emulsifying properties in the digestive system, reacting with fats and oils as a detergent to form smaller particles of the consumed compounds. In addition, the same amphipathic properties that allow bile acids to emulsify using lipids also make them membrane-disruptive mediums.
Sources
- http://www.elmhurst.edu/~chm/vchembook/558detergent.html. Elmhurst College. 2003.
- http://chemistry.about.com/od/howthingswork/f/detergentfaq.htm. About.com. Anne Marie Helmenstine, Ph.D. 2009
- http://www.chemistry.co.nz/surfactants.htm. LA & HA Campbell. 1999
General Information
[edit | edit source]
Also, referred to as terpenoids or prenol lipids, isoprenoids are any of a class of organic compounds composed of two or more units of hydrocarbons, with each unit consisting of five carbon atoms arranged in a specific pattern. These compounds are derived from five-carbon isoprene units and are biosynthesized from a common intermediate known as mevalonic acid, which is itself synthesized from acetyl-CoA. These lipids are considered to be the largest group of natural products, playing a wide variety of roles in physiological processes of plants and animals, and having a number of commercial uses.
In living organisms, these compounds range in function from pigmentation and fragrances to vitamins and precursors of sex hormones. Uses in industrial settings range from flavorings, solvents and raw materials for chemicals. These compounds are more commonly associated with being the main ingredients of perfumes and incense. But these are just a few of the many commercial uses for isoprenoids, which to this day are still expanding.
Isoprenoid Isolation and Identification
[edit | edit source]Isolation of isoprenoids from their natural sources is achieved through numerous procedures. Substances that are volatile and plentiful, such as turpentine, are best obtained by distillation of oleoresins. Rosin acids and fatty acids, which occur together in tall oil, are separated by fractional distillation at reduced pressure. Other compounds that are more on the rare side are best isolated by chromatography. A very laborious process known as enfleurage is employed for the isolation of heat-sensitive perfume ingredients. This process involves carefully placing the petals containing the oily isoprenoids in thin layers of purified fat to dissolve the oils. The oils are then recovered from the fat by washing the solution with alcohol.
Physical properties of an isoprenoid compound are the main factor that must be looked at to identify the best purification technique that will yield the best results. A heat-sensitive compound will not be efficiently and successfully purified with techniques such as distillation or sublimination, which require heat. Similar compounds in one source are best isolated and purified by treatment into a mixture of new substances that can be more readily separated. After separation, the original products can be attained by reconverting them. Generally, solid compounds can be purified by recrystallization and volatile compounds (solid or liquid) are best purified by distillation. Similar and nonvolatile substances might require chromatography to best achieve separation.

The determination of the elemental composition of isoprenoids is not a difficult task due to their simple hydrocarbon makeup and the availability of simple and reliable procedures for quantitative analysis of carbon and hydrogens. The presence of the other element in these compounds, oxygen, does not interfere with the analysis of the carbon and hydrogen makeup in a significant way, but does make it a more laborious task. A particularly difficult compound that was difficult to determine throughout the history of isoprenoid studies was camphor, which had a total of more than 30 different structures presented before the correct one was found.
Structures are more commonly identified by using nuclear magnetic resonance (NMR). This technique uses the help of a magnetic field to generate a response of the compound to this energy. This data is then collected and interpreted (computer analysis gives the best results to this day). Mass spectrometry, x-rays, and IR spectroscopy present other alternatives to NMR. High-resolution mass spectrometry enables the exact chemical formula of a compound to be determined, while X-ray crystallography permits the detailed spatial location of each atom to be determined from a diffraction pattern of the crystallized form of the structure.
Structural Features and Some Isoprenoid Compounds
[edit | edit source]The five-carbon unit that constitutes the basic building block of isoprenoids is a hydrocarbon called isoprene. This compound is a branched-chain unsaturated hydrocarbon, meaning it has one or more double bonds between carbon atoms. Isoprenoids can have one or more functional chemical groups attached to their carbon backbone, such as hydroxyls and carbonyls, which make up the diversity of isoprenoids. Isoprenoids can be classified as monoterpenes (C10H16) sesquiterpenes (C15H24), diterpenes (C20H32), triterpenes (C30H48), tetraterpenes (C40H64) or other polyterpenes (C5H8)n . Many isoprenoids are arranged in a “head to tail” manner in which a carbon atom of one isoprene unit is bonded to a carbon atom of the next isoprene unit. Triterpenes and tetraterpenes, though, will tend to bond its isoprene units in a “tail-to-tail” fashion.

Monoterpenes (C10H16)
[edit | edit source]The first isoprenoids that were studied initially in the history of these lipids were the monoterpenes, the molecules of which contain 10 carbon atoms. These compounds are isolated from their natural sources by distillation of the plant matter with steam. They are characterized as volatile oils that are less dense than water and have normal boiling points in the range of 150 to 185ᵒC. Fractional distillation at reduced pressures work best at purifying these compounds. Some widely known monoterpene derivatives include the oxygenated acyclic monoterpene derivative citronellol and its corresponding aldehyde citronellal, which occur in oil of citronella. Others include the compound citral, which is found in lemongrass oil and geraniol, most commonly found in Turkish geranium oil.

The process used to convert citronellal is widely used commercially to supplement the natural sources of menthol, since this process produces a mixture of stereoisomeric menthols by catalytic hydrogenation. Citral is used in the production of rose-scented perfumes. The reduction of citral with sodium amalgam yields a compound called geraniol, which is responsible for the scent. Also, citral can be treated with acetone to condense it and yield pseudoionone, which is in turn treated with acid to produce β-ionone. Although not considered a terpene, β-ionone is commonly used as a starting material for the synthesis of vitamin A.
Sesquiterpenes (C15H24)
[edit | edit source]Sesquiterpenes are of lower volatility than monoterpenes and thus, can be isolated from their natural sources by extraction. Purification of these compounds involves either vacuum fractional distillation or chromatography. The oxygenated sesquiterpenes are the most commonly encountered ones. Sesquiterpenes may be acyclic or contain rings. A longer chain length and more double bonds contributes to a wide variety of cyclization structures. For example, there are two arrangements of isoprene units found in bicyclic sesquiterpenes: that of the cadalene types and the eudalene types. An example of each form is cadinene, the principal component of the optically active oils of cubeb and cade, which is of the cadalene type and β-Selinene, which is present in celery oil, and is of the eudalene type.


Diterpenes (C20H32)
[edit | edit source]Diterpenes are known to be antimicrobial and antiinflammatory. They derive from geranygeranyl pyrophosphate. One well known diterpene is phythol, an oxygenated acyclic diterpene that is the building block of the chlorophyll molecule. This compound is obtained on treatment with an alkali solution. Phythol is similar to vitamin A in terms of arrangement of their isoprene units (head-to-tail arrangement).

Another common diterpene is the commercially utilized tricyclic abietic acid. This compound is a carboxylic acid that constitutes the major portion of rosin. Rosin is the residue left after the isolation of turpentine and is the nonvolatile portion of the oleoresin of members of the pine family. It is used in the production of varnish and coating materials, among many other products. The versatility of this compound was discovered due to it being one of the cheapest organic acids available.

Triterpenes (C30H48)
[edit | edit source]An example of a triterpene is the acyclic hydrocarbon squalene (pictured above). This constitutes more than half of the liver oil of certain species of sharks and is otherwise, widely distributed in nature. Among shark liver oil, is it found in other fish liver oils, vegetable oils, fungi, and in human earwax. Squalene is a metabolic intermediate in the biosynthesis of cholesterol.
The most common types of triterpenes found in nature are those having five carbon rings. β-amyrin has usually been associated with much of the research done in the study of triterpenes. β-amyrin is found in the resin elemi, which is obtained primarily from trees in the family of flowering plants known as Burseraceae. Its skeletal structure shares many similarities with that of squalene and cholesterol.

Tetraterpenes (C40H64)
[edit | edit source]Some biologically important tetraterpenes include carotenoids, which account for the yellow, orange or red fat-soluble plant and animal pigments. Although they have eight lesser hydrogen atoms than the general formula of tetraterpenes, they are still considered tetraterpenes since they can be built up from isoprene units. Lycopene is the red pigment of the ripe tomato. It's structure has an interruption to the more common head-to-tail bonding of isoprene units. This interruption contains a single tail-to-tail attachment, which gives lycopene its symmetrical structure. Many tetraterpenes generally have this feature.

β-carotene is another more common tetraterpene, which is the yellow pigment of the carrot. This compound is a precursor to vitamin A, which means it is of nutritional importance since animals are able to cleave this compound at the point of symmetry to promote the production of active vitamin A. This vitamin is of considerable importance in the synthesis of pigments in the eye that are necessary for healthy vision.

Polyterpenes (C5H8)n
[edit | edit source]A well-known example of a polyterpene is rubber, in which n = 4,000-5,000. Oxidative degradation followed by x-ray diffraction revealed rubber to be made up of repeating units. The vulcanization of such a compound involves cross-linking between the chains through sulfur atom (disulfide bridges).

Biosynthesis
[edit | edit source]The synthesis of isoprenoids begins with acetyl coenzyme A, a compound derived from acetic acid and coenzyme A. In the first part of the process, mevalonic acid and isopentenyl pyrophosphate (IPPP) occur as important intermediates.

Afterwards, the formation of geranyl pyrophosphate (GPP, precursor of monoterpenes), occurs by the transformation of an IPPP to dimethylallyl pyrophosphate (DMAPP). DMAPP then combines with an IPPP to form this precursor.

Further combinations of the pyrophosphate compounds in a similar fashion can yield precursors to larger polyterpenes, such as the formation of squalene from GPP and IPPP, which can then form such isoprenoids as Lanosterol.
Overview
[edit | edit source]Researchers have learned much of what they know about membranes by constructing artificial membranes in the laboratory. In artificial membranes, different lipids separate from each other based on their physical properties, forming small islands called lipid rafts. Thus, Lipid rafts are possible island like structure present in cellular membranes.
These rafts have a higher concentration of certain specialized lipids, called glycosphingolipids, and cholesterol than do non-raft parts of the membrane. Rafts also are distinguished by a different assortment of proteins. Certain types of proteins cluster together in rafts, while others remain mostly outside of rafts. The big question is, to what extent do these rafts, seen readily in artificial membranes, actually exist in living cells? Using advanced laboratory methods and imaging techniques, some researchers found evidence that rafts, indeed, do form in living cellular membranes, but these rafts may be small and transitory. Although the existence of lipid rafts in cellular membranes remains controversial, many scientists believe they serve as communication hubs by recruiting proteins that need to come together in order to transmit a signal. Researchers are beginning to link lipid rafts with a variety of diseases, including AIDS, Alzheimer’s, anthrax, and atherosclerosis.
Structural Details
[edit | edit source]The amount and types of proteins and lipids found in membranes can vary depending on the membrane's location in the body or which organelle it surrounds. These differences allow a wide variety of processes to be performed with membranes. In the fluid mosaic model, lipids and proteins exhibit lateral mobility and are constantly changing positions. Cholesterol and sphingolipids, which include both sphingomyelin and glycosphingolipids, are also commonly found in membranes. It has been assumed that individual proteins and lipids move independently and randomly, but some patches of aggregation have been observed in membranes, too. Relatively new studies using artificial membranes have shown that certain proteins and lipid patches tend to aggregate with each other to form island-like LIPID RAFTS. These island-like lipid rafts can be analogous to Jello with embedded fruits where the fruits play the role as proteins. Lipid rafts have a noticeably higher concentration of cholesterol and glycosphingolipids than other areas of the membrane. Certain types of proteins are also more likely to be found in rafts than others. Specifically, an example of a protein found in lipid rafts are acylated proteins, and some have proposed that the reaction between cholesterol and their unsaturated acyl chains promotes the formation of lipid rafts. Also, proteins have been associated with lipid rafts via glycosylphosphatidylinositol (GPI) anchors, which covalently attach proteins at the membrane's external service. There is still question as to the existence of lipid rafts in living cells, so the current theory is that the lipid rafts likely do exist but are small and transitory.
Sphingolipids
[edit | edit source]Examples of common sphingolipids include sphingosine and sphingosine 1-phosphate. They are crucial in regulating many cellular processes—such as migration, proliferation, differentiation, and immune responses. These sphingolipids are central for cellular signaling, more knowingly familiar as a bioactive lipid mediator.
Recent evidences have proven that the signaling function of these sphingolipids indicate association of apoptosis and growth arrest. Sphingosine is capable of directly inhibiting protein kinase C, along with other effects on various protein kinases in vitro. Additionally, sphingosines have shown signs of regulating the pro-survival adaptor protein 14-3-3 by directly interacting with it and enabling the phosphorylation and inactivation of the adaptor component via protein kinases. Examples of this are shown in PKA and PKC-gamma. By doing so, the pro-survival pathway is completely barricaded.
Another common sphingolipid is ceramide, which like sphingosine, is associated with apoptosis. Ceramide correlated with growth arrest and apoptosis through levels of various responses of stress stimuli. Ceramide demonstrates modulation of proteins either through direct catalytic activities, or by facilitation of subcellular localization.
As a result, regulation of sphingosine kinase activity has illustrated crucial cellular functionality. Monitoring of such kinases allowed the determination of cell fate, whether a cell will survive or proliferate. Additionally, studies of sphingosine kinases have assisted in the control of adhesion, differentiation, and immune reaction and have increased possibilities of providing powerful information for therapeutic actions.
Types and Functions of Lipid Rafts
[edit | edit source]Two types of lipid rafts have been proposed: planar lipid rafts, also called glycolipid rafts, and caveolae. Planar rafts are continuous with the plane of the plasma membrane and do not have distinctive morphological features. Caveolae, on the other hand, are flask shaped inward foldings of the plasma membrane that contain caveolin proteins, which are a group of proteins involved in receptor-independent endocytosis.
It has been proposed that lipid rafts serve to collect proteins when needed for signal transmissions and are possibly linked to diseases like AIDS. The ability to recruit signaling proteins and molecules into their island-like microdomains would give lipid rafts the possibility to play an important role in neurotransmitter signaling. The hypothesis for this is that the environment of lipid rafts promote kinetically favorable interactions required for signal transduction. On the other hand, they could also potentially serve to separate signaling molecules and decrease signals by inhibiting the interactions. The caveolin types of lipid rafts could possibly be involved with endocytosis and cholesterol transportation, too.
Controversy about Lipid Rafts
[edit | edit source]One of the primary reasons for the controversy over lipid rafts has stemmed from the challenges of studying lipid rafts in living cells. Lipid rafts are small microdomains ranging from 10–200 nm in size, which is below the classical diffraction limit of a light microscope. This means lipid rafts are quite difficult to visualize, let alone observe in action. Also, experimental studies of lipid rafts have led many to believe that they are transitory and do not remain intact for very long. An argument against the useful existence of lipid rafts in living cells is that the time scale of their actual existence is unknown. If lipid rafts exist, they may only occur on a rapid time scale that is too short to serve much purpose and therefore irrelevant to biological processes.
References
[edit | edit source]1. http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=&_xRefDocId=pii%23b012443710900363x&_user=4429&_fmt=high&_orig=na&_cdi=20141&_refWorkId=264&view=c&_acct=C000059602&_version=1&_urlVersion=0&_userid=4429&md5=22bb48e7aeabf3a57722193092abfd22, Science Direct, Encyclopedia of Biological Chemistry, Pg. 584-587, 12/2/2009
2. Inside the Cell, U.S. Department of Health and Human Services, 12/2/2009
3. http://www.uoguelph.ca/~fsharom/research/gpi.html
4. http://www.bms.ed.ac.uk/research/others/smaciver/Cyto-Topics/lipid_rafts_and_the_cytoskeleton.htm
- ↑ U.S. Department of Health and Human Services. Inside the Cell. September 2005.<http://www.nigms.nih.gov>.
Metabolomics is the study of nonproteinaceous, low molecular weight intermediates (metabolome) in the cell and the fats (lipids) constitute the largest subset in the metabolome. Hence, lipidomics is a subcategory of metabolomics and it is the detailed analysis and characterization of the structure and function of lipids. There are two types of lipidomics: targeted and untargeted. Targeted lipidomics is where the lipid species to be monitored is known before starting the analysis and untargeted lipidomics is simply exploring and trying to find new lipids. For untargeted lipidomics, mass spectrometry is used and searched for new m/z ratio peaks. A large variety of lipids constitute the cellular membranes, and analyzing their interactions with membrane-associating proteins can lead to further analysis, such as insight on drug interactions.
Sir JJ Thomson invented the mass spectrometer, which observes the mass-to-charge ratio of the sample. According to the article, "Applications of Mass Spectrometry to Lipids and Membranes," there are four main applications of mass spectrometry to the field of lipidomics (1): 1) Two current mass spectrometry-based lipidomics approaches 2) Mass spec-based structure determination of novel lipids 3) Deuterium exchange mass spectrometry to study the location and orientation of proteins associated with lipid membranes 4) Advances in the imaging of lipids in tissue through Matrix Assisted Laser Desorption mass spectrometry (1)
The LIPIDS MAPS Consortium has defined 8 categories of lipids based on their chemically function backbone: fatty acyls, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides
There are two methods to create sample preparations for mass spectrometry. The first method is comprehensive lipidomics analysis by separation simplification, which is better known as CLASS. This method is based on separation of different lipid categories using extraction and chromotographic separation. First, the components of the lipid are chromatographically separated and then added directly into the mass spectrometry ("divide-and-conquer" approach). To separate the components of the fatty acids, there are two methods, high powered liquid chromotagraphy and gas chromotography. HPLC couples directly into the mass spectrometer and "gas chromatography requires derivataization of the free fatty acid" since no electrospray ionization is in it (1). Electrospray ionization allows for the ionization of proteins in a liquid medium without derivitization. A small amount of ammonium acetate can be added to the electrospray ionization solution in order to increase the ionization efficiency. The CLASS approach minimizes ion suppression observed in the other method, shotgun approach. Shotgun approach omits chromatographic separation and analyzes all the lipid classes together while employing different ion source polarities. For this method, the researcher must carefully select electrospray ionization, electrospray ionization additives, and mass spectrometry mode. The downfall to this method is ion suppression. The low level species are not detected; therefore, the CLASS approach is a more preferred method.
For both methods, a general cell or tissue preparation is needed (1): 1) Cells or tissues cultured and subjected to a probe (activation or perturbation) 2) Lipidome is compared to a control unperturbed sample 3) Lipids stored within cell wall need to be disturbed through sonication and produce a uniform homogenate 4) Prior to extraction, use internal standard to enable absolute quantity of lipids in the sample 5) Solvent containing extracts evaporated and remaining lipids resuspended in liquid medium optimal for direct infusion into the mass spectrometer or in a medium compatible with either gas or liquid chromatography prior to CLASS (1)
References
[edit | edit source]1. Annu Rev Biochem. 2011 Jun 7;80:301-25. Applications of mass spectrometry to lipids and membranes. Harkewicz R, Dennis EA. Source Department of Chemistry and Biochemistry and Department of Pharmacology, School of Medicine, University of California at San Diego, La Jolla, California 92093-0601, USA. rharkewicz@ucsd.edu Targeted versus Untargeted Lipidomics (Assays) "Targeted" assay is where the lipid species to be analyzed is already labeled and known before the analysis process. An example is multiple reaction monitoring method, which has "excellent sensitivity and is ideal for accurate quantitation" (1). Through this method, the researcher only finds what he is looking for.
"Untargeted" assay is more of a discover as you research type of assay of lipids. An example of this assay is using the mass spectrometer in full-scan mode, which searches for new mass-to-charge ratio peaks. This method has assumptions such as that the species will ionize efficiently and that the mass spectrometer is sensitive. Furthermore, after a new, novel lipid is discovered, further investigation and research can be conducted on the newly observed lipid. There is a concern/question that arises while conducting untargeted assays, such as are biologically significant lipid species being overlooked?
There are many examples of novel, unexpected lipids, such as the discovery of the concept of fatty acids by Michael Eugene Chevreul. He was the first lipid specialist to discover the concept of fatty acids and also discovered cholesterol and glycerol. Other examples of novel lipids include the "novel family of N-acylphosphoserine derivatives in the brain of a mouse and also glycoerophosphocholine in retina of the eye" (1).
Reference
[edit | edit source]1. Annu Rev Biochem. 2011 Jun 7;80:301-25. Applications of mass spectrometry to lipids and membranes. Membranes must be able to separate or come together in order for the cells to absorb, transport, and release molecules. Receptor-mediated endocytosis is a common process in which cells take up molecules. In many cases, a large protein complex initially binds to a receptor located on the surface of the cell. Once the complex is bound to the receptor, specific proteins cause the membrane to invaginate. This ultimately causes the membrane to break off and fuse to form a vesicle.
This process is apparent when dealing with cholesterol in the blood (LDL). LDL binds to a specific integral membrane protein, LDL receptor. The LDL-LDL-receptor complex invaginates, therefore "breaking" the membrane. The LDL-LDL-receptor complex also separates such that LDL separates from the LDL recptor. The formed vesicle containing LDL fuses with a lysosome, which results in the degradation of LDL and the release of a cholesterol molecule. Biosynthesis of triacylglycerol starts with the acylation of the two free hydroxyl groups of L-glycerol 3-phosphate by two molecules of fatty acyl-CoA to produce phosphatic acid. This is then hydrolyzed by phosphatidic acid phosphatase to form a 1,2-diacylglycerol. 1,2-diacylglycerol is then converted to triacylglycerol by transesterification with a third fatty acyl-CoA. [1]
In humans, hormones can affect the biosynthesis of triacylglycerol. Insulin is one example of a hormone that promotes the conversion of carbohydrates to triacylglycerols. [1]
- ↑ a b Cox, Michael M. and Nelson, David L. Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. Print.
Biosynthesis of triacylglycerol starts with the acylation of the two free hydroxyl groups of L-glycerol 3-phosphate by two molecules of fatty acyl-CoA to produce phosphatic acid. This is then hydrolyzed by phosphatidic acid phosphatase to form a 1,2-diacylglycerol. 1,2-diacylglycerol is then converted to triacylglycerol by transesterification with a third fatty acyl-CoA. [1]
In humans, hormones can affect the biosynthesis of triacylglycerol. Insulin is one example of a hormone that promotes the conversion of carbohydrates to triacylglycerols. [1]
- ↑ a b Cox, Michael M. and Nelson, David L. Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. Print.
Lipids regulate the amplitude, duration, and subcellular location of signaling by lipid second messenger responsive kinases. Generally, this activation is regulated by membrane targeting modules that regulate the function of kinase domains within the same polypeptide. Protein kinase C (PKC) acts as the primary lipid-regulated kinase, providing a prototype for lipid-controlled kinase activation that is followed by kinases throughout the kinome, such as Akt (Protein Kinase B).
Cellular membranes form a platform of intense signaling activity. Acting as the site where extracellular signals are first received by the cell, they not only recruit and activate effector molecules, but they also provide an initiation to the activated effector molecules throughout the cell. Protein kinases possess an extremely common class of effector molecules that transduce signals, coming from the plasma membrane. These kinases can be found in the plasma membrane, illustrated by the tyrosine kinase growth factor receptors, or can be either soluble or amphipathic membrane proteins that translocate on and off cellular membranes in response to appropriate signals. Soluble proteins are found by membranes through protein scaffolds, but there are amphipathic membrane kinases whose members directly bind lipid second messengers through specific membrane targeting modules.
For PKC, The interaction of the membrane-targeting modules on the membrane, with high affinity, produces a conformational change that releases an auto-inhibitory pseudosubstrate segment from the substrate-binding cavity, allowing substrate binding and downstream signaling. This active conformation depends on lipid binding. In the case of Akt, engaging the PH domain on membranes serves the purpose of allowing priming phosphorylations of Akt. Once phosphorylated at two key positions, Akt is locked in an active conformation, and, unlike PKC, activity is independent of lipid binding. On the other hand, activity is regulated by the phosphorylation state of Akt. Therefore, lipids control the activity of a phosphorylated PKC, an event that dissociates Akt activity from lipid binding.
Lipid second messengers relay an abundance of signals that control cell growth and survival. Direct binding of protein kinases to these lipid second messengers serves as a first step to transduce information into the cell. The activity of these kinases is regulated and deregulation of this activity results in pathological states such as cancer.
Source: http://newtonlab.ucsd.edu/documents/Newtonreview2009.pdf
Unsaturated and Odd-Chain Fatty Acids Require Additional Steps for Degradation
[edit | edit source]Most fatty acids have an even number of carbon atoms due to the process by which they were synthesized, thus the beta-oxidation pathway can successfully complete the degradation of these molecules. However, there are more steps required for fatty acid chains that are not as simple, i.e. those that are unsaturated or which have an odd number of carbons.
To oxidize unsaturated fatty acids, an isomerase and a reductase are required
[edit | edit source]Unsaturated fatty acids, while prominent in our diets, are more complicated to metabolize than saturated ones. In addition to the reactions required for the degradation of saturated fatty acids, the degradation of unsaturated fatty acids calls for two supplementary enzymes: an isomerase and a reductase.
For example, let’s look at the oxidation of the unsaturated fatty acid palmitoleate (pictured), which has 16 carbons with one double bond between C9 and C10. Just like saturated fatty acids, this unsaturated fatty acid is first activated then transported across the inner membrane of the mitochondria.
The now palmitoleoyl CoA proceeds to undergo 3 degradation cycles carried out by the same enzymes that oxidize saturated fatty acids. A problem arises however when the cis-Δ3-enoyl CoA is formed in the third round of these degradations: cis-Δ3-enoyl CoA is not a substrate for acyl CoA dehydrogenase. As seen in the picture, there is a double bond between C3 and C4 which prevents a double bond from forming between C2 and C3. This obstacle in degradation is overcome by shifting the position and configuration of the cis-Δ3 double bond to a trans-Δ2 double bond; this new reaction is facilitated by cis-Δ3-Enoyl CoA isomerase. Now that the double bond is between C2 and C3, the rest of the reactions relevant to saturated fatty acid oxidation can be done on trans-Δ2-enoyl CoA.
Excess polyunsaturated fatty acids (ones with more than one double bond) are degraded via beta-oxidation and are important to humans as precursors for signal molecules. There is another obstacle to be overcome when dealing with polyunsaturated fatty acids, however, which can be discerned by looking at the oxidation of the 18-carbon polyunsaturated fatty acid linoleate (pictured). Linoleate has cis-Δ9 and cis-Δ12 double bonds; when the cis-Δ3 double bond is formed after 3 rounds of beta-oxidation, it is converted into a trans-Δ2 double bond by the same isomerase mentioned in the palmitoleate degradation. After another round of beta-oxidation, the acyl CoA produced contains a cis-Δ4 double bond. When this species is dehydrogenated by acyl CoA dehydrogenase it yields a 2,4-dienoyl intermediate.

This intermediate is not a substrate for the next enzyme in the beta-oxidation pathway, so 2,4-dienol CoA reductase is employed to convert the intermediate into trans-Δ3-enoyl CoA. 2,4-dienol CoA reductase does this by using NADPH to reduce the 2,4-dienoyl intermediate to trans-Δ3-enoyl CoA. cis-Δ3-enoyl CoA isomerase can then convert the trans-Δ3 into the trans-Δ2 form, which is an acceptable intermediate in the beta-oxidation pathway.
To sum up: odd-numbered double bonds are taken care of by the isomerase while even-numbered double bonds are handled by the isomerase and the reductase together.
In the final thiolysis step, odd-chain fatty acids yield propionyl CoA
[edit | edit source]Fatty acids with an odd number of carbons are a minor species and are oxidized in the same way as fatty acids with an even number of carbons. The difference is that when the odd-numbered fatty acid is oxidized, it produces propionyl CoA and acetyl CoA in the final round of degradation rather than two molecules of acetyl CoA. The activated 3 carbon unit in propionyl CoA, once converted into succinyl CoA, enters the citric acid cycle.

The pathway that takes propionyl CoA to succinyl CoA requires vitamin B12 for a certain rearrangement. The conversion of propionyl CoA to succinyl CoA is pictured. The carboxylation reaction is catalyzed by propionyl CoA carboxylase, which is a biotin enzyme with a catalytic mechanism analogous to that of pyruvate carboxylase.
Vitamin B12 contains a cobalt atom and a corrin ring
[edit | edit source]
Cobalamin enzymes catalyze 3 types of reactions: intramolecular rearrangements, methylations, and the reduction of ribonucleotides to deoxyribonucleotides. The two reactions in mammals that require coenzyme B12 are: (1) the conversion of L-methylmalonyl CoA into succinyl CoA and (2) methylation of homocysteine to form methionine. Reaction (2) is particularly important as Met is necessary for the generation of coenxymes that play a role in the synthesis of purines and thymine. The basic structure of a cobalamin is pictured: the core consists of a corrin ring with a central cobalt atom.
The rearrangement in the formation of succinyl CoA is catalyzed by methylmalonyl CoA (mechanism)
[edit | edit source]
During this rearragement, two groups attached to adjacent carbon atoms are excahnged, the process being catalyzed by coenzyme B12. The first step in these rearrangements is the carbon-cobalt bond of 5’-deoxyadenosyl being cleaved (a homolytic cleavage reaction). This creates the Co2+ coenzyme form and a radical of 5’-deoxyadenosyl (pictured).

The highly reactive radical serves its purpose by abstracting a hydrogen atom from the substrate, forming 5’-deoxyadenosine and a radical substrate, which spontaneously rearranges (the carbonyl CoA group travels to the position previously occupied by the neighboring carbon atom’s hydrogen). This produces a different radical, which abstracts an H atom from the methyl group of 5’-deoxyadenosine thus completing the rearrangement. To sum up, coenzyme B12’s function in these intramolecular migrations is to be a source of free radicals for the abstraction of H atoms.
Peroxisomes are also sites for fatty acid oxidation
[edit | edit source]While the majority of fatty acid oxidation takes place in the mitochondria, some oxidation can occur in peroxisomes (a small membrane-bound organelle found in most eukaryotes). One of the main roles of oxidation that takes place in the peroxisomes is the oxidation of fatty acids down to octanoyl CoA (a better substrate for beta-oxidation in mitochondria). The difference between beta-oxidation in the peroxisomes versus the mitochondria is found in the initial dehydrogenation reaction. Instead of capturing high-energy electrons as FADH2 for the electron transport chain as in mitochondrial oxidation, in peroxisomal oxidation the flavoprotein acyl CoA dehydrogenase transfers electrons from the substrate to FADH2 then to oxygen, yielding hydrogen peroxide. In order to degrade H2O2 into water and oxygen there is a large concentration of the enzyme catalase within the peroxisomes.
When fat breakdown predominates, ketone bodies are formed
[edit | edit source]If the fat and carbohydrate degradation in a cell are balanced properly the acetyl CoA from fatty acid oxidation will enter the citric acid cycle. For entry into the citric acid cycle to be granted to the acetyl CoA, it must combine with the oxaloacetate; the concentration of available oxaloacetate, normally formed from pyruvate (the product of glycolysis of glucose), is dependent on the supply of carbohydrate.
When oxaloacetate isn’t readily available (as with those who suffer form diabetes), acetyl CoA is diverted to the formation of D-3-hydroxybutyrate and acetoacetate, which are often called ketone bodies.
Acetyl CoA forms into acetoacetate in 3 steps (pictured), and the first step is catalyzed by thiolase. The overall reaction is 2Acetyl CoA + H2O --> acetoacetate + 2CoA + H+
D-3-hydroxybutyrate is formed when acetoacetate in the matrix of the mitochondria is reduced by D-3-hydroxybutyrate dehydrogenase. Acetoacetate undergoes an additional slow, spontaneous decarboxylation to acetone because it is a beta-ketoacid.
In some tissues ketone bodies are used as a major fuel source
[edit | edit source]D-3-hydroxybutyrate is formed when acetoacetate in the matrix of the mitochondria is reduced by D-3-hydroxybutyrate dehydrogenase. Acetoacetate undergoes an additional slow, spontaneous decarboxylation to acetone because it is a beta-ketoacid.
The conversion of acetoacetate into acetyl CoA occurs in 2 steps. First the transfer of CoA from succinyl CoA to acetoacetate activates the molecule. This step is catalyzed by a CoA transferase. In the second step, thiolase cleaves acetoacetyl CoA to yield two molecules of acetyl CoA, which can then enter the citric acid cycle. When 3-hydroxybutyrate is reacted to generate acetyl CoA, an additional step is required: it must be initially oxidized by NAD+ to produce acetoacetate.
To sum up, we can look at ketone bodes as water-soluble, easily transportable forms of acetyl units. Having high levels of acetoacetate is a sign of abundant levels of acetyl units; this leads to a decrease in the rate of lipolysis.
Liposomes are artificially constructed vesicles consisting of a phospholipid bilayer. First discovered in 1961 by Alec Bangham, a British scientist studying blood clotting, liposomes are now being studied for their potential in both laboratory techniques as well as medical applications. Of particular interest are their ability to cross cell membranes and to transport certain types of drugs to pre-designated locations within the human body.

Structure
[edit | edit source]Liposomes are spherical structures, usually between 15nm and 1000nm in diameter. Various targeting ligands can be attached to their surface to direct them to the appropriate sites within cells; these include, but are not limited to, membrane proteins. It is important to differentiate liposomes from micelles; even though both of these macromolecular complexes are spherical and consist of lipids, a micelle is normally formed from ionized fatty acids, whereas a liposome consists of phospholipids. Furthermore, micelles consist of only a single layer of lipids, with their non-polar carbon tails clustered together at the center (therefore not allowing any water soluble compounds on the interior), whereas liposomes are constructed from a bilayer that does allow charged molecules on the inside. This is due to the presence of the hydrophilic glycerol-phosphate-alcohol heads of phospholipids, which define both the outer and inner surfaces of liposomes.
Composition of Liposomes
[edit | edit source]The Major Structural Components of Liposomes are: [1]
- Phospholipids - Phospholipids are the main component of the liposome's membrane. The phospholipids used in liposomes are further categorized into natural and synthetic phospholipids. The most common phospholipid used is known as lecithin (also known as phosphatidylocholine) and is amphipathic.
- Cholestorol - Cholesterol molecules in the membrane increases separation between choline head groups which reduces the normal hydrogen bonding and electrostatic interaction.
Classification of Liposomes
[edit | edit source]
Liposomes can be classified into several types according to their following features:[2]
- Size
- Number of Lamellae
| Liposome Types | Size | Number of Lamellae |
|---|---|---|
| Small Unilamellar Vesicles (SUV) | 20 nm - 100 nm | Single |
| Large Unilamellar Vesicles (LUV) | 100 nm - 400 nm | Single |
| Giant Unilamellar Vesicles (GUV) | 1 µm and Larger | Single |
| Large Multilamellar Vesicles (MLV) | 200 nm - ~3 µm | Multiple |
| Multivesicular Vesicles (MVV) | 200 nm - ~3 µm | Multiple |
- Note: Lamellae refers to the lipid bilayers.
- Note: Refer to Diagram to Right for Better Idea.
Synthesis
[edit | edit source]Liposomes are frequently synthesized by mixing and dissolving the phospholipids in organic solvent, such as chloroform or a chloroform-methanol mixture. A clear lipid film is subsequently formed by removal of the solvent, and hydration of this film eventually leads to formation of large, multilamellar vesicles (LMVs). An LMV consists of more than one bilayer, creating a complex the structure of which has several layers, analogous to the structure of an onion. Each bilayer is separated from the next by water. Smaller liposomes are produced by disrupting LMVs using sonication (agitation by sound-waves). This process yields small, unilamellar liposomes (SUVs) between 15nm and 50nm in diameter. These are not very stable and tend to form larger vesicles. Storing them above their phase transition temperature can help prevent formation of those larger vesicles. In order to synthesize larger liposomes, the method of extrusion is commonly used. Following several freeze-thaw cycles, the lipid suspension (the LMV suspension) is forced through polycarbonate filters containing pores, which leads to formation of liposomes with diameters similar to the size of the pores. This technique, if employed with pores of approximately 100nm in diameter, allows for the formation of large, unilamellar vesicles (LUVs) approximately 120nm – 140nm in size. These sizes are significantly more reproducible than those achieved through sonication.
Scientific Use
[edit | edit source]The study of phospholipid bilayers can help clarify several of their characteristics, such as their permeability under varying pH and temperature, their fluidity, and their electro conductivity. This is usually done with sheets of bilayers, which can be more easily generated and are more stable, not vesicles such as liposomes. However, liposomes have been found useful in studies of phase transitions and lattice spacing and were thus used for such purposes in the late 1960s and early 1970s.
Pharmaceutical and Medical Applications
[edit | edit source]The greatest potential of liposomes lies in the medical field, where their ability to deliver drugs and other compounds to specific areas of an organism are under active investigation. The basis for this ability is that the hydrophilic compounds contained by a liposome cannot pass through the hydrophobic core of the lipid bilayer, and are thus trapped on the inside. The attachment of glycosylated membrane proteins to the outside or the vesicle can help direct the liposome to the desired cells. These binding ligands may also be responsible for the fusion of the vesicle with the diseased cell. This process of drug delivery often lessens the toxicity of the drugs, which are sheltered from interaction with other non-target cells. Additionally, this mode of delivery can be more efficient, as long circulating liposomes may accumulate in a region of higher than average blood circulation, such as an inflammation site, a tumor, or other diseased areas.
Factors other than the membrane protein mediated fusion of vesicles to cellular membranes can contribute to the release of the compound contained in the liposome. An example of this is the pH-triggered permeability change of a heterogeneous liposome. Liposomes under certain pH conditions can become “leaky” and thereby release the compounds contained within. Doxorubicin, a cancer drug, is delivered to tumor cells by this mechanism, which does not affect overall liposome stability.
Advantages of Liposomes in Drug Delivery
[edit | edit source]- Since the 1960's Liposomes and their use in the medicinal field has been greatly explored by pharmaceutical companies. Liposomes have many advantages as a method of drug delivery. These advantages are as follows:
- Liposomes are biocompatible, completely biodegradable, non-toxic, flexible, and nonimmunogenic.
- Liposomes have both a lipophilic and aqueous environment making it useful for delivering hydrophobic, amphipathic, and hydrophilic medicines.
- Liposomes with their layers encapsulates the drug and serves as a protection of the drug from the environment as well as acting as a sustained release mechanism. This encapsulation also serves to protect sensitive areas from the drug as well.
- Liposomes are extremely versatile in the form which they may be administered. These forms include suspension, aerosol, gel, cream, lotion, and powder which can then by administered through most common routes of medicinal administration.
- Liposomes are also flexible in their size, and as such they can enclose a wide size range of molecules.
- Liposomes can aide with active targeting as it has flexibility in coupling with site-specific ligands.
- [4]
Disadvantages of Liposomes in Drug Delivery
[edit | edit source]- Despite all the wonderful advantages, Liposomes do have some disadvantages when compared with other methods of drug delivery.
-
- Liposomes encapsulated drugs require a high production cost.
- Liposomes may have leakage and fusion of encapsulated drugs.
- The liposome phospholipid may undergo oxidation and hydrolysis.
- Liposomes have a shorter half-life.
- Liposomes have lower solubility.
- [5]

List of Drugs Using Liposomes
[edit | edit source]| Marketed Drug | Targeted Disease |
|---|---|
| DoxilTM | Kaposi’s Sarcoma |
| DaunoXomeTM | Kaposi’s Sarcoma, Breast & Lung Cancer |
| AmphotecTM | Leishmaniasis, Fungal Infections |
| Fungizone® | Leishmaniasis, Fungal Infections |
| VENTUSTM | Inflammatory Diseases |
| ALECTM | Expanding lung diseases in babies |
| Topex-Br | Asthma |
| Depocyt | Cancer therapy |
| Novasome® | Smallpox |
| Avian Retrovirus Vaccine | Chicken pox |
| Epaxal –Berna Vaccine | Hepatitis A |
| Doxil® | Refractory Ovarian Cancer |
| NyotranTM | Fungal Infections |
References
[edit | edit source]- 1. http://www.nanopharmaceuticals.org/Liposomes.html
- 2. http://www.fasebj.org/content/24/5/1308.full
- 3. http://www.ncbi.nlm.nih.gov/pubmed/17092599
- 4. http://www.avantilipids.com/index.php?option=com_content&view=article&id=1384&Itemid=372
- 5. http://www.bioportfolio.com/resources/pmarticle/264214/Heterogeneous-Liposome-Membranes-With-Ph-triggered-Permeability-Enhance-The-In-Vitro-Antitumor.html
- 6. D Papahadjapoulos and N Miller."Phospholipid Model Membranes I. Structural characteristics of hydrated liquid crystals." Biochimica et Biophysica Acta. 135. (1967) 624-638.
- 7. H Trauble and D H Haynes."The volume change in lipid bilayer lamellae at the crystalline-liquid crystalline phase transition." Chem. Phys. Lipids. 7. (1971) 324-335.
- 8. Berg, Jeremy (2012). Biochemistry. New York: W. H. Freeman and Company. ISBN 978-1-4292-2936-4.
Liposome Classifications References
[edit | edit source]- ↑ "Liposome: A versatile platform for targeted delivery of drugs." Shri B. M. Shah College of Pharmaceutical. Sanjay S. Patel (M. Pharm), 2006.
- ↑ ‘Galenic Principles of Modern Skin Care Products’, Professor Rolf Daniels, 2005
- ↑ "Liposome Drug Products: Chemistry Manufacturing and Control Issues." Arthur B. Shaw, Ph.D, 2001.
- ↑ "Liposome: A versatile platform for targeted delivery of drugs." Shri B. M. Shah College of Pharmaceutical. Sanjay S. Patel (M. Pharm), 2006.
- ↑ "Liposome: A versatile platform for targeted delivery of drugs." Shri B. M. Shah College of Pharmaceutical. Sanjay S. Patel (M. Pharm), 2006.
- ↑ "Liposome: A versatile platform for targeted delivery of drugs." Shri B. M. Shah College of Pharmaceutical. Sanjay S. Patel (M. Pharm), 2006.
Definition
[edit | edit source]Lipid droplets are the lipid storage organelles of all organisms. Their important roles include cellular and organismic energy storage[1] Lipid droplets could be found nearly in every cell. They are cytoplasmic organelles that store lipids; some examples are triglycerides and cholesterol. Under nutrient deprivation, droplet triglycerides are hydrolyzed to create free fatty acids, which are then oxidized and used to provide energy.
Lipid droplets in a cellular organism are typically composed of nonpolar, hydrophobic lipids, also known as neutral lipids. The droplets contain a hydrophobic center that is encircled by a phospholipid monolayer. There are two major types of lipid droplets; sterol esters and triacylglycerols. As the lipid droplets form there can be a variety of lipid droplet proteins along the monolayer. The amount of lipid droplets in each cell can vary and currently scientists are unable create an algorithm to predict the lipid droplet concentration. [2]
Lipid droplets are formed on or in close proximity of endoplasmic reticulum. Through the use of the electron microscope, it was possible to see that lipid droplets and cisternae have a close relationship but the mechanism of formation has yet to be discovered. 2
There are over 800 genes that affect lipid droplet accumulation. When testing to see how these multitudes of genes affect lipid droplet formation, knockdown of genes led to differing lipid droplet formation changes. Depending on the gene that was knocked-down, some of the lipid droplets decreased in concentration, increased in size, or increased in size and were more dispersed. Proteins were also found to affect lipid droplet formation. For example, with the over-expression of fat-inducing transcript-2 (FIT2) proteins, there are more lipid droplets. Consequently, knockdown of FIT2 leads to fewer lipid droplets. 2
Lipid droplets are capable of interacting with other cellular organelles. They interact with the endoplasmic reticulum, endosomes, mitochondria, and peroxisomes. These interactions can sometimes mediate some cellular mechanisms such as lipid trafficking, neutral lipid metabolism, and synthesis/catabolism of steroid hormones. A majority of these interactions occur at the endoplasmic reticulum.2

http://gladstoneinstitutes.org/u/rfarese/research_pics/lipid_droplet.jpg
Properties of Lipid Droplets
[edit | edit source]1. Almost every cell has lipid droplets or has the capacity to produce them
2. Some bacteria use lipid droplets to store lipids themselves
3. Lipid droplets' abundance varies greatly in cells.
4. Lipid droplets' size also varies greatly depending on cell types. Many cells are known to contain small LDs (100-200 nm). On the other hand, in white adipocytes, for instance, have diameters up to 100 micrometers, occupying most of the cytoplasm.
Functions of Lipid Droplets
[edit | edit source]1. Generally they are intracellular lipid reservoirs that become useful as they provide building blocks for cell membranes or substrates for energy metabolism[3]
2. Lipid droplets may be used as places for synthesizing some lipids. For instance, TGs are produced in the Er and lipid droplets.
3. Lipids droplets may be used to store some proteins.
4. Lipid droplets are involved in hepatitis C virus assembly [4]
Lipid Droplets and Tissues
[edit | edit source]Adipose Tissue In mammals and birds, adipose tissues is located in specific areas in the body and are regulated by hormones. They are more prominently used as insulation for endotherms. There are two types of adipocytes: white adipocytes and brown adipocytes. Adipocytes are the cells that are utilized by the body to store lipids in lipid droplets. White adipocytes store lipids in a single large lipid droplet in the cytoplasm. They are also known to store cholesterol esters and fat-soluble vitamins. The white adipocytes utilize leptin to regulate the lipid storage. Brown adipocytes catabolize lipids in order to generate heat. They store lipids in smaller and more numerous lipid droplets in the cytoplasm.
Liver The liver has the second greatest lipid in LD form storage capacity. They store the lipid droplets in hepatocytes. In humans, a high amount of lipid droplets can lead to an illness called fatty liver.
Small Intestine The small intestine is where a majority of nutrient absorption occurs. By utilizing its microvilli, the small intestine is able to absorb 95% of a meal's fat content. The lipid droplets are stored in intestinal enterocytes. The enterocytes have a large surface area and are able to store and synthesize triacylglycerols(TGs).
Yolk Sac The yolk sac stores lipids in a similar manner to intestinal enterocytes and liver hepatocytes. They are able to store lipid droplets and export via apoB-containing lipoproteins.
Skeletal Muscle Muscle is not known to be a large lipid storage unit. When humans store lipids in their skeletal muscles it is usually a side effect of obesity because of insulin resistance. There is an exception with this in highly-trained athletes. Athletes use a high volume of ATP when they are training so the body needed a way to keep a lipid reserve for when the muscles need more energy. This compensation occurs with the lipid droplets being stored in skeletal muscle cells near the mitochondria. This is considered the athelete's paradox because lipid droplet storage in skeletal muscles is usually a bad sign.
Adrenal Cortex The adrenal cortex is able to store large amounts of sterol esters(SEs). The SEs are most likely stored in the form of cholesterols for steroid hormone synthesis. Also the yellow coloring of the tissue can be credited to the lipids.2
Reference
[edit | edit source]1. Nature.com. Nature Publishing Group, n.d. Web. 07 Dec. 2012.
2. http://www.ncbi.nlm.nih.gov/pubmed/22524315
- ↑ http://www3.mpibpc.mpg.de/groups/jaeckle/pages/Project_Beller/project_Beller.html
- ↑ http://www.ncbi.nlm.nih.gov/pubmed/22524315
- ↑ “Lipid Droplets and Cellular Lipid Metabolism” Annual Review of Biochemistry Vol 81 587-714, by Tobias C. Walther and Robert V. Farese Jr
- ↑ “Lipid Droplets and Cellular Lipid Metabolism” Annual Review of Biochemistry Vol 81 587-714, by Tobias C. Walther and Robert V. Farese Jr