Structural Biochemistry/Enzyme
Enzymes are macromolecules that help accelerate (catalyze) chemical reactions in biological systems. This is usually done by accelerating reactions by lowering the transition state or decreasing the activation energy.

Some biological reactions in the absence of enzymes may be as much as a million times slower. Virtually all enzymes are proteins, though the converse is not true and other molecules such as RNA can also catalyze reactions. The most remarkable characteristics of enzymes are their ability to accelerate chemical reactions and their specificity for a particular substrate. Enzymes take advantage of the full range of intermolecular forces (van der waals interactions, polar interactions, hydrophobic interactions and hydrogen bonding) to bring substrates together in most optimal orientation so that reaction will occur. Also, enzymes can be inhibited by specific molecules by called competitive, uncompetitive, and noncompetitive inhibitors.
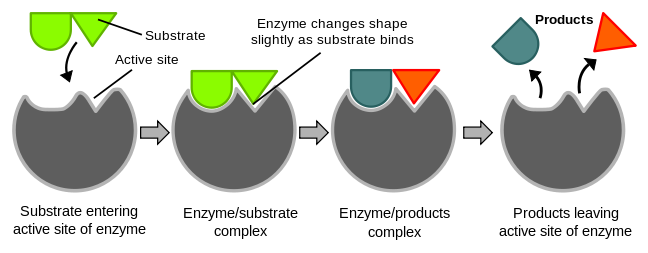
Catalysis happens at the active site of the enzyme. It contains the residues that directly participate in the making and breaking of bonds. These residues are called the catalytic groups. Although enzymes differ widely in structure, specificity, and mode of catalysis a number of generalizations concerning their active sites can be made:
1. The active site is a three dimensional cleft or crevice formed by groups that come from different parts of the amino acid sequence - residues far apart in the amino acid sequence may interact more strongly than adjacent residues in the sequence.
2. The active site takes up a relatively small part of the total volume of an enzyme. Most of the amino acid residues in an enzyme are not in contact with the substrate, which raises the question of why enzymes are so big. Nearly all enzymes are made up of more than 100 amino acid residues. The "extra" amino acids serve as a scaffold to create the three dimensional active site from the amino acids that are far apart in the primary structure. In many proteins the remaining amino acids also constitute regulatory sites, sites of interaction with other proteins, or channels to bring the substrate to the active sites.
3. Active sites are unique microenvironments. In all enzymes of known structure, substrate molecules are bound to a cleft or crevice. Water is usually excluded unless it is a reactant. The nonpolar microenvironment of the cleft enhances the binding of substrates as well as catalysis. Nevertheless, the cleft may also contain polar residues. Certain of these polar residues acquire special properties essential for substrate binding or catalyis.
4. Substrates are bound to enzymes by multiple weak interactions. Stated above
5. The specificity of binding depends on the precise defined arrangement of atoms in the active site. Because the enzyme and the substrate interact by means of short-range forces that require close contact, a substrate must have a matching shape to fit into the site. However, the active site of some enzymes assume a shape that is complementary to that of the substrate only after the substrate is bound. This process of dynamic recognition is called induced fit.
Enzymes are highly specific and may require cofactors for catalysis. A cofactor is a non-protein chemical compound bound to a protein; there are 2 types of cofactors: Metals and organic/metalloorganic (which are derived from vitamins). An example of a metal cofactor is zinc and the enzyme, carbonic anhydrase, tightly binds the zinc at the active site. The process involves binding water to carbon dioxide and deprotonating it into carbonic acid. Then the carbonic acid becomes a bicarbonate ion due to the displacement of water.
Catalysts can fasten the reaction speed by lowering the activation energy (not the transition state) of the process. The active site is a location on the enzyme which has complementary shape to the substrate. It is also where the amino acids with a complementary charge, polarity and shape to the ligand are.
The enzyme function and catalysis result from the ability to stabilize the transition state in a chemical reaction. The transition state is the highest energy species in a reaction. It is a transitory molecular structure that is no longer the substrate but is not yet the product. It is the most seldom occupied species along the reaction pathway. The difference in free energy between the transition state and the substrate is called the Gibbs free energy of activation or simply the activation energy.
Thus we can see the key to how enzymes operate: Enzymes accelerate reactions by decreasing the activation energy. The combination of substrate and enzyme creates a reaction pathway whose transition state is lower than that of the reaction in the absence of the enzyme. Because the activation energy is lower more substrate molecules have the energy required to reach the transition state.
It is important to note that enzymes have evolved specifically to recognize the transition states of chemical reactions. Therefore, enzymes do not bind to any reactive species before the species have actually begun to react; enzymes only recognize and bind the transition states of such species. In fact, if enzymes were to bind to the reactants of a reaction "on sight", or immediately, this would result in an even higher activation energy than before! For this reason, enzymes recognize only the transition state and bind to reactive species only when this high-energy state has been achieved. The fact that enzymes can recognize structures as specific and short-lived as transition states is a testament to their incredible specificity and efficiency.
Each enzyme is optimized for a particular reaction transition state. This ensures that enzymes will not compete with each other and hinder cellular reactions instead of help them. Enzyme inhibition occurs when the activity of a given enzyme is disrupted or interrupted in some fashion. Inhibitors can be molecules that have a similar shape, structure, or charge to the substrate in question so that the active site of an enzyme will "mistake" the inhibitor for the substrate. This affects the affinity of the enzyme for the substrate, as well as the rate of the overall reaction. Several types of inhibition can occur in the cell; more detailed explanations on these can be found in the corresponding sections.
Because of the active sites, enzymes are highly specific catalysts. These catalysts are governed by the ability to lower the free energy of thermodynamics to overcome transition states. The Michaelis-Menten Model describes the kinetic properties of many enzymes.
The interaction between the substrate and the enzyme helps accelerate the reaction, and the specificity of enzymes result in minimal side reactions.
It is of great importance to note that an enzyme cannot alter the laws of thermodynamics and consequently cannot alter the equilibrium of the reaction. The amount of product formed for a reaction utilizing an enzyme is always equal to the amount of product form of the same reaction occurring in the same reaction mixture without the enzyme. The enzyme just allows the reaction to reach its equilibrium faster. The equilibrium position is a function only of the free-energy difference between reactants and products.
6. Enzymes only alter reaction rate, not the reaction equilibrium. Enzyme cannot alter the laws of thermodynamics; therefore, it cannot alter the equilibrium of a chemical reaction. Enzyme is present, the amount of products form faster compared with enzyme is absent. Enzyme is only accelerating the reaction rate, not shipping the position of equilibrium (free energy, delta G)
Lock and Key Model
[edit | edit source]The "lock and key" model was first proposed by an organic chemist named Emil Fischer in 1894. In this model, the "lock" refers to an enzyme and the "key" refers to its complementary substrate. Each enzyme has a highly specific geometric shape that is complementary to its substrate. In order to activate an enzyme, its substrate must first bind to the active site on the enzyme. Only then will a catalytic reaction take place. However, like a lock and a key, the enzyme and substrate shape must be complementary and fit perfectly. Designed by evolution the active site for enzymes is generally highly specific in its substrate recognition and has the ability to distinguish between stereoisomers.
Induced Fit
[edit | edit source]According to the Lock and Key Model, the geometric shape of both enzymes and substrates can not be changed as they are both predetermined. Thus, the binding of the substrate to the enzymes active site does not alter the shape of the enzyme. While this theory helped explain the specificity of the enzyme, it does not explain the stability of the transition state for it would require more energy to reach the transition state complex. Thus the induced fit model was proposed in which enzymes like proteins are flexible. The concept of induced fit is that when a substrate binds to the active site of an enzyme, there is a conformational change and structural adaptation that makes this binding site more complementary and tighter. In essence the substrate does not simply bind to a rigid active site but instead the macromolecules, weak interaction forces, and hydrophobic characteristics on the enzyme surface mold into a precise formation so that there is an induced fit where the enzyme can perform maximum catalytic function.
Transition State Theory
[edit | edit source]
Transition state theory states that in an enzyme catalysis, the enzyme binds more strongly to its "transition state complex rather than its ground state reactants." In essence, the transition state is more stable. The stabilization of the transition state lowers the activation barrier between reactants and products thus increasing the rate of reaction or enzymatic activity as this will favor the increase of formation of the transition state complex.
In the transition state theory, the mechanism of interaction of reactants is irrelevant. However, the colliding molecules that take place in the reaction must have sufficient amount of kinetic energy to overcome the activation energy barrier in order to react. In many cases, temperature, pH, or enzymes can be changed to facilitate the stabilization of the transition state as well as statistically increasing the probability for molecules colliding and forming the transition state complex. For a bimolecular reaction such as Sn2, a transition state is formed when the two molecules’ old bonds are weakened and new bonds begin to form or the old bonds break first to form the transition state and then the new bonds form after. The theory suggests that as reactant molecules approach each other closely they are momentarily in a less stable state than either the reactants or the products.
Methods
[edit | edit source]- Some catalysts provide a charge to a molecule to make it more attractive to other reactants. Acids are an example for this kind of catalyst. They give the reacting species a positive charge to attract the negative or partially negative reactant, increasing the chance for the two species to collide and react.
- Some catalysts increase the local concentration of reactants so that they are more likely to collide.
- Some catalysts may modify the shape of one reactant to be more susceptible to other molecule.
Enzymatic Strategies and Examples
[edit | edit source]1. Covalent Catalysis - Through the course of catalysis, a powerful nucleophile is temporarily attached to a part of the substrate. The nucleophile is contained in the active site. A proteolytic enzyme chymotrypsin is an excellent example of this strategy. It is a substrate forming a transient covalent bond with residues in the active site or with a cofaster, which adds additional intermediate and reduce the energy of later transition.
2. General Acid/Base Catalysis - Water often acts as a donor or acceptor, but in Acid/base catalysis, the molecule which donates or accepts a proton is NOT water. This strategy incorporates base and acid catalysis to shorten reaction times. In the case of Chymotrypsin, the enzyme uses a histidine residue as a base catalyst to enhance the nucloephilicity of serine analogous to how histidine residue in carbonic anhydrase facilitates the removal of a proton from a zinc bound water molecule to yield hydroxide.
3. Catalysis by approximation - In this method, reactions favored by bringing together the two substrates to a single binding surface on enzymes. The two substrates are brought together to one area and this increases the rate of the reaction. NMP kinase for example, brings tow nucleotides together to improve the transferring of phosphoryl groups.
4. Metal Ion Catalysis - Metal ions can be involved as a catalyst in many different ways. Zinc can help the formation of a nucleophile. It makes the pka of water change from approximately 14 to 7, which allows it to be protonated at neutral pH. It can also stabilize negative charges by acting as an electrophile in a complex. Metal ions are also used to increase the binding energy of substrates, holding them together. A metal ion may also serve as a bridge between the enzyme and substrate acting as a cofactor in cases of NMP kinases.
Catalytic Mechanisms
[edit | edit source]1. Proteases (chymotrypsin and trypsin): are any enzyme that conducts proteolysis (protein catabolism) by hydrolysis of the peptide bonds linking amino acids together in the polypeptide chain.
Sample Experiment: Site-Directed Mutagenesis Applying Polymerase Chain Reaction (PCR) & Oligonucleotide Primers that contains the desired mutation in a newly synthesize strand, engineering a mis-match during first cycle DNA can develop a mutation.
2. Carbonic Anhydrase (metalloenzymes) These enzymes catalyzes the rapid interconversion of carbon dioxide and water to bicarbonate and protons, a reversible reaction that occurs rather slowly in the absence of catalyst.
3. Restriction Endonucleases (BamHI) It is a restriction enzyme that cleaves double stranded DNA at specific recognition nucleotide sequences (restriction site).
4. Nucleaside Monophosphate Kinases (NMP Kinase) These enzymes transfer phosphate groups from high energy donor molecule (ATP) to specific substrates (phosphorylation).
Enzyme's Cofactors for Activity
[edit | edit source]
The catalytic activity of enzymes depends on the presence of small molecules called cofactors. The role of the catalytic activity varies with the enzyme and its cofactors. In general, those cofactors can execute chemical reactions which cannot be performed by the standard 20 amino acids. An enzyme without cofactor is called apoenzyme, however the one with completely catalytically active is called holoenzyme.
Cofactors can be divided into two individual groups: Metal and Coenzymes. Metals are important for enzymes because they are molecular assistants that play a vital role in some of the enzymatic reactions that fuel the body metabolism. They also act to stabilize the shapes of enzymes. For example, iron helps the protein hemoglobin transport oxygen to organs in the body and copper helps superoxide dismutase in sopping up dangerous free radicals that accumulate inside the cells. Coenzymes are small organic molecules that often derived from vitamins. Coenzymes can be either tightly or loosely bound to the enzyme. Tightly bound ones are called prosthetic groups, while loosely bound coenzymes are like substrates and products, bind to the enzyme and get released from it. Enzymes that use the same coenzymes often perform catalysis by the similar mechanisms.
Enzyme Classification
[edit | edit source]| Class | Type of Reduction | Examples |
|---|---|---|
| Hydrolases | Catalyze hydrolysis reactions | Estrases Digestive enzymes |
| Isomerases | Catalyze isomerization (changing of a molecule into its isomer) | Phospho hexo isomerase, Fumarase |
| Ligases | Catalyze bond formation coupled with ATP hydrolysis. | Citric acid synthetase |
| Lyases | Catalyze a group elimination in order to form double bonds (or a ring structure). | Decarboxylases Aldolases |
| Oxidoreductases | Catalyze oxidation-reduction reactions | Dehydrogenases Oxidases |
| Transferases | Catalyze the transfer of functional groups among molecules. | Transaminase Kinases |
The classification of an enzyme is shown within the table as it's class and the type of reduction the enzyme goes through. An example of a name is glucose phosphotransferase. In this reaction ATP transfers one of its phosphates to glucose: ATP + D-glucose -> ADP + D-glucose 6-phosphate. Since this process "transfers" a phosphate group to glucose, it is within the classification of transferases, hence the name "glucose phosphotransferase." Since many enzymes have common names that do not refer to their function or what kind of reaction they catalyze, a enzyme classification system was established. There are six classes of enzymes that were created with subclasses based on what they catalyze so that enzymes could easily be named. Depending on the type of reaction catalyzed, an enzyme can have various names. These classes are Oxidoreductases, Transferases, Hydrolases, Lyases, Isomerases, and Ligases. This is the internation classification used for enzymes. For example, a common oxidoreductase is dehydrogenase. Dehydrogenase is known as an enzyme that oxidizes a substrate and transferring protons. Enzymes are normally used for catalyzing the transfer of functional groups, electrons, or atoms. Since this is the case, they are assigned names by the type of reaction they catalyze. This allowed for the addition of a four-digit number that would precede EC(Enzyme Commission) and each enzyme could be identified. The reaction that an enzyme catalyzes must be known before it can be classified.
Oxidoreductases catalyze oxidation-reduction reactions where electrons are transferred. These electrons are usually in the form of hydride ions or hydrogen atoms. When a substrate is being oxidized it is the hydrogen donor. The most common name used is a dehydrogenase and sometimes reductase will be used. An oxidase is referred to when the oxygen atom is the acceptor.
Transferases catalyze group transfer reactions. The transfer occurs from one molecule that will be the donor to another molecule that will be the acceptor. Most of the time, the donor is a cofactor that is charged with the group about to be transferred.
Hydrolases catalyze reactions that involve hydrolysis. This cases usually involves the transfer of functional groups to water. When the hydrolase acts on amide, glycosyl, peptide, ester, or other bonds, they not only catalyze the hydrolytic removal of a group from the substrate but also a transfer of the group to an acceptor compound. These enzymes could also be classified under transferase since hydrolysis can be viewed as a transfer of a functional group to water as an acceptor. However, as the acceptor's reaction with water was discovered very early, it's considered the main function of the enzyme which allows it to fall under this classification.
Lyases catalyze reactions where functional groups are added to break double bonds in molecules or the reverse where double bonds are formed by the removal of functional groups.
Isomerases catalyze reactions that transfer functional groups within a molecule so that isomeric forms are produced. These enzymes allow for structural or geometric changes within a compound. Sometime the interconverstion is carried out by an intramolecular oxidoreduction. In this case, one molecule is both the hydrogen acceptor and donor, so there's no oxidized product. The lack of a oxidized product is the reason this enzyme falls under this classification. The subclasses are created under this category by the type of isomerism.
Ligases are used in catalysis where two substrates are litigated and the formation of carbon-carbon, carbon-sulfide, carbon-nitrogen, and carbon-oxygen bonds due to condensation reactions. These reactions are couple to the cleavage of ATP.
Kinetics
[edit | edit source]The study of the rates of chemical reactions is called kinetics, and the study of the rates of enzyme-catalyzed reactions is called enzyme kinetics. A kinetic description of enzyme activity will help us understand how enzymes function. For example, the rate V is the quantity of A that disappears in a specified unit of time. It is equal to the rate of the appearance of P, or the quantity of P that appears in a specified unit of time.
A -----> P
The rate V is the quantity of A that disappears in a specified unit of time. It is equal to the rate of the appearance of P, or the quantity of P that appears In a specified unit of time.
V = ∆A/∆T = ∆P/∆T
If A is yellow and P is colorless, we can follow the decrease in the concentration of A by measuring the decrease in the intensity of yellow color with time. Consider only the change in the concentration of A for now. The rate of the reaction is directly related to the concentration of A by a proportionality constant, k, called the rate constant.
V = k[A]
The Michaelis-Menten Model
[edit | edit source]The Michaelis-Menten model is used to describe the kinetic properties of many enzymes. In this model, an enzyme(E)combines with a substrate(S)to form an enzyme-substrate(ES)complex, and proceed to form a product(P)or to dissociate into E and S.
The rate of formation of product,V0, can be calculated by the Michaelis-Menten equation:
Vmax is the reaction rate when the enzyme is completely saturated with substrate. KM is the Michaelis constant, which is the substrate concentration at the half of the maximum reaction rate. The kinetic constant kcat is called the turnover number, which is the number of substrate molecules converted into product per unit time at a single catalytic site when the enzyme is saturated with substrate. It often count for most enzyme between 1 and 104per second.
Allosteric enzymes is an important class of enzymes. Its catalytic activity can be regulated. It has multiple active sites which display co-operativity, as evidenced by a sigmoidal dependence of reaction velocity on substrate concentration. We also find that K max is the substrate concentration in which the overall reaction rate at that particular time is half of V max. V max on the other hand, is the maximum reaction rate in which the active site is completely saturated with substrate. As a result of this physical characteristic, we see that no matter how much substrate is consequently added, the relative rate of the reaction remains unchanged as additional substrate do not contribute to any kinetic interaction with binding the active site. The affinity also eventually does not change as more substrate is increased and the reaction goes towards equilibrium.
Replicative DNA polymerase
[edit | edit source]There have been studies of the three multi-subunit DNA polymerase enzymes in the nucleus. This provides insights into the mechanism of the replication machinery in eukaryotic cells. The first DNA polymerase structure to be solved by crystallography was the Klenow fragment of E. coli DNA polymerase I. This crystallization revealed a structure that was likened to the palm, fingers, and thumb of a right hand. Studies of the Klenow fragment showed that DNA was bound within the cleft and that the fingers and thumb architecture is conserved in many of the polymerase families. The polymerase active site residues are located in the palm domain. The fingers are important for nucleotide binding, and the thumb domain binds the DNA.

Conclusion Enzymes speed up the reaction. They can increase the reaction rates by a factor of 10^6 or more. Some of the enzymes require the cofactors for activity. The cofactors include vitamin-derived organic molecules (coenzymes), and metal ions. The enzymes' other tasks include decreasing the free energy of activation of chemical reactions. The first step in catalysis is forming an enzyme substrate complex. Besides, the substrates are recognized by enzymes is accompanied by conformational changes at active sites.
Temperature affect on catalytic activity of enzymes
[edit | edit source]In general as one increases the temperature the catalytic activity of an enzyme increases, which is the classical model and understanding of temperature affect on catalytic activity. Though, the classical model has been discovered to be a questionable depiction of how temperature really affects the catalytic activity of enzymes. The new model, the equilibrium model, has been studied by Roy M. Daniel and Micahel J. Danson and gives a new understanding of how temperature affects the catalytic activity of enzymes.
Classical Model
The classical model depicts the effects of temperature on an enzyme. Mainly depicts that as one increases temperatures the catalytic rate increases exponentially and the amount of active enzyme will decrease because of denaturization. The classical model does not show if there is an optimal temperature at which, catalytic activity is at the max.
Equation representing the classical model: Vmax= kcat[E0]e-kinactt
Equilibrium Model
In the equilibrium model, it introduces an inactive form of the enzyme that is reversible to be active. The new factor of reversibility between active and inactive form of enzymes is temperature dependent where the amount of enzyme is.
The equation describing the amount of active enzyme at any point, [Eact] = ([E0]-[X])/( 1+ Keq)
In this context inactive enzyme does not mean the enzyme is denatured, all it means is that the active site has been changed just enough to the point where the enzyme can not bind to the substrate, hence it is reversible.
The model shows an optimal temperature where enzyme activity would be the greatest, which is different from the classical model. It takes into account the variation Vmax with time and temperature and also includes the factor of four parameters; G*cat, G*inact, Heq and Teq. This model is currently used to best describe the evolution of the enzyme’s active site due to temperature and also best explain the effect of temperature on enzymes.
Key difference between the two models
The classical model is temperature dependent and doesn’t depict an optimal temperature for catalytic activity.
References
[edit | edit source]
Biochemistry 7Ed, by Jeremy M. Berg, John L. Tymoczko, and Lubert Stryer. 2007.
Berg, Jeremy M., Tymoczko, John L., Stryer, Lubert. Biochemistry. Seventh Edition.
http://www.ncbi.nlm.nih.gov/pubmed/20554446
http://ars.els-cdn.com/content/image/1-s2.0-S0968000410000897-gr1.jpg



![{\displaystyle {\frac {1}{v_{0}}}={\frac {K_{M}+[S]}{V_{\max[}S]}}={\frac {K_{M}}{V_{\max }}}\cdot {\frac {1}{[S]}}+{\frac {1}{V_{\max }}}.}](https://wikimedia.org/api/rest_v1/media/math/render/svg/3f2e414bdbcccae96840f4a5c32e303e33b5be72)