Structural Biochemistry/Organic Chemistry/Proteins
Protein molecules contain polypeptide chains made from sequences of the 20 amino acids. These amino acids are linked together by a peptide bond that is formed by condensation of two amino acids with the elimination of the elements of water. Protein function is dependent on its tertiary structure. Proteins tend to fold into three- dimensional structures because of the sequence of amino acids. Proteins also contain functional groups from each amino acid. These groups are reactive and also contribute to protein function. Proteins also interact with one another and with other macromolecules. Proteins can be rigid or flexible. This allows certain proteins to be found in different parts of the cell such as the cytoskeleton or in soft tissue.
Importance of Proteins
[edit | edit source]Enzymes are proteins that catalyze chemical reactions. Enzymes speed up the reactions in biological systems by lowering the activation barrier needed to start that reaction.
Hormones are proteins that are chemical messengers in the body. These proteins are sent to different parts of the body to send or receive messages. Hormones are very important in regulating the human body and keeping the body in a state of homeostasis. Some protein hormones include insulin, growth hormone, Luteinizing hormone (LH), follicle-stimulating hormone (FSH), and thyroid-stimulating hormone (TSH). These proteins are part of the glycoprotein hormones.
Transport Proteins are also used to transportation. For example, hemoglobin is a metalloprotein(protein that contain a metal as cofactor) that transports an oxygen in the red blood cells with the help of iron.
Motor Proteins help convert chemical energy to mechanical energy which relates to muscular motion in an organism. Examples are actin and myosin.
Protective Proteins protect cells by releasing, making antibodies, fighting and destroying foreign objects. Antibodies are gamma globulin proteins.
Structural Proteins help maintain the structure of a variety of biological components like cells and tissues in an organism. Collagen, elastin, α-keratin, sklerotin[check spelling], and fibroin are all examples of proteins that contribute to the formation of an organism's body.
Storage Proteins that contain energy and can be digested during metabolism of the organism. Examples are egg ovalbumin and milk casein.
Membrane Proteins include receptors and membrane transport. The receptors in the membrane of cells allow ions to pass through. These prevent unwanted objects from coming into the cell. These receptors also determine if the cell is excited to create an action potential or not. Membrane transport is important because it allows ions, proteins, and other macromolecules to pass through the cell membrane.
Classification of proteins by location
[edit | edit source]External proteins-proteins outside of cells and are found in multicell organisms.
Internal proteins-proteins that are inside cells and perform functions for intercellular needs.
Membrane proteins-proteins that are embedded in the bilayer of the membrane of on the edges of the membrane helping with intracellular interactions.
Virus proteins-usually the coat for viruses
Enzymes
[edit | edit source]Enzymes are proteins that speed up the reaction rate. Many reactions cannot occur without the use of an enzyme.

This graph shows how the presence of an enzyme lowers the activation energy and therefore speeds up the reaction. Reactions with enzymes can speed up to 10 billion times faster than those without an enzyme. The rate at which the enzyme works is affected by the substrate and enzyme concentration, temperature, and pH.
Amino Acids
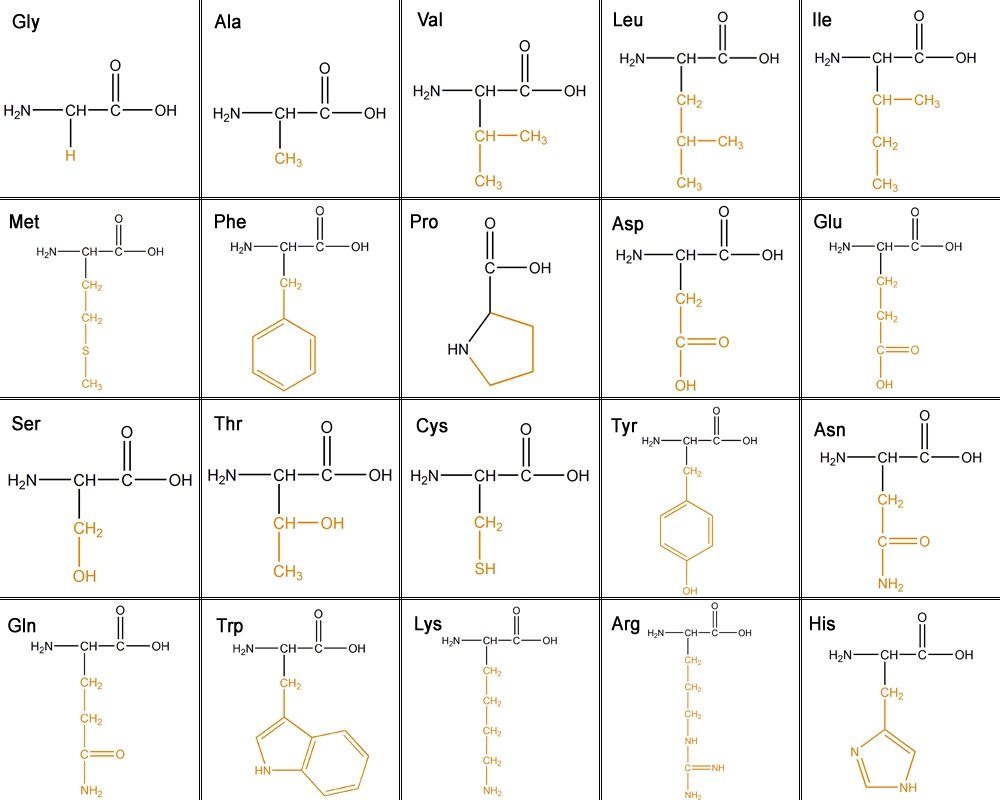
[edit | edit source]There are 20 amino acids that make up proteins. The main chain has an N terminal which is an amino group (NH2) and a C terminal which is a carboxyl group (COOH). The side chains make each amino acid unique.
The 20 different amino acids can be classified into six different classes based on their side chain(R group).
1. Aliphatic - carbon side chain. The longer the aliphatic chain, the more hydrophobic.
Glycine, Alanine, Valine, Leucine, Isoleucine
2. Hydroxyl or Sulfur containing- The OH is reactive, hydrophilic (water loving), polar and uncharged. Sulfur is very reactive.
Serine, Threonine, Cysteine, Methionine
3. Cyclic- Proline
4. Aromatic-
Phenylalanine - purely hydrophobic
Tyrosine - OH reactive
Tryptophan - less hydrophobic due to its NH groups
5. Basic- hydrophilic and positively charged.
Lysine, Arginine, Histidine
6. Acidic and their amide- negatively charged.
Aspartate, Glutamate, Asparagine, Glutamine
This is the general structure of an amino acid in its unionized form.
This table shows all the amino acids and the their side chains. From left to right they are: glycine(Gly), alanine(Ala), valine(Val), Leucine(Leu), isoleucine(ILe), methionine(Met), phenylalanine(Phe), proline(Pro), aspartic acid(Asp), glutamic acid(Glu),serine(Ser), threonine(Thr), cystine(Cys), tyrosine(Tyr), asparagine(Asn), glutamine(Gln), tryptophan(Trp), lysine(Lys), arginine(Arg), and histidine (His).
Proteins are made from many amino acids. They are connected together by peptide bond. Peptide bonds are formed by condensation, the loss of awater molecule and between the carboxyl group with the aminogroup.
The reaction above shows how two alanine are linked together by a peptide bond. The bond is formed between the n terminal amino group and the c terminal carboxyl group. Two hydrogens and an oxygen come out in this reaction which produces water. The peptide bond acts almost like a double bond due to the resonance of the carbonyl. Because of this, there is no rotation about this bond so therefore conformation is limited. This limits stereochemistry in that almost all peptide bonds in proteins are trans isomers to limit steric hindrance between the R groups. Only proline can be either cis or trans because the energy levels these two isomers show are about the same(the side chain of proline has similar distance to the adjacent R group in either isomer). The reason for that is because proline's side chain form a ring with the alpha-amino group. Proline is the only amino acid whose side chain form a ring with the alpha-amino group.
Structures
Proteins can fold into four different structures. These structures determine protein function and characteristics.
Primary Structure- The primary structure of a polypeptide is its amino acid sequence, from beginning to end. The primary structures of polypeptides are determined by genes. Genes carry the information to make polypeptides with a defined amino acid sequence. For the protein to function correctly, each amino acid needs to be in order as the genes assigned. Even a little change in the amino acid sequence would affect the shape of the protein and its ability to function. An average polypeptide is about 300 amino acids in length, and some genes encode polypeptides that are a few thousand amino acids long.
Secondary Structures- The amino acid sequence of a polypeptide, together with the laws of chemistry and physics, cause a polypeptide to fold into a more compact structure. Amino acids can rotate around bonds within a protein. This is the reason proteins are flexible and can fold into a member of shapes. Folding can be irregular or certain regions can gave a repeating folding pattern. Such repeating patterns are called secondary structures. The two types are the α-helix and β-sheet. In an α-helix, the polypeptide backbone forms a repeating helical structure that is stabilized by hydrogen bonds. These hydrogen bonds occur at regular intervals and cause the polypeptide backbone to form a helix. In a β-sheet, regions of the polypeptide backbone come to lie parallel to each other. When these regions form hydrogen bonds, the polypeptide backbone form a repeating zigzag shape called a β-sheet.
Tertiary Structure- As the secondary structure becomes established due to the primary structure, a polypeptide folds and refolds upon itself to assume a complex three-dimensional shape called the protein tertiary structure. The tertiary structure is the three-dimensional shape of a single polypeptide. It's usually a result of interactions among the R groups of the amino acids that make up the polypeptide. For some proteins, such as ribonuclease, the tertiary structure is the final structure of a functional protein. Other proteins are composed of two or more polypeptides and adopt a quaternary structure. Tertiary structure is important in regards to enzymatic activity.
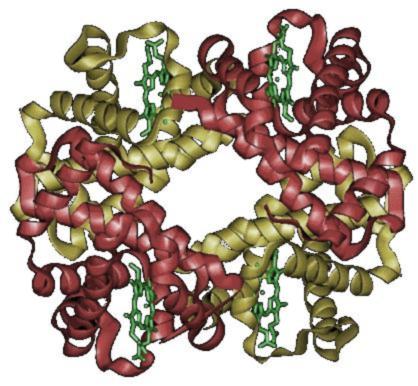
Quaternary Structure- Most functional proteins are composed of two or polypeptide that each adopt a tertiary structure and then assemble with each other. The individual polypeptides are called protein subunits. Subunits may be identical polypeptides or they may be different. Each subunit has a non-protein component that is necessary for the protein to function correctly. These components are called heme. When proteins consist of more than one polypeptide chain, they are said to have quaternary structure and are also known as multimeric proteins, meaning many parts.
Factors that influence protein structure
[edit | edit source]Several factors determine the way that polypeptides adopt their secondary, tertiary and quaternary structures. The amino acid sequences of polypeptides are the defining features that distinguish the structure of one protein from another. As polypeptides are synthesized in a cell, they fold into secondary and tertiary structures, which assemble into quaternary structures for most proteins. As mentioned, the laws of Chemistry and physics, together with amino acid sequence, govern this process. Five factors are critical for protein folding and stability:
2. Ionic bonds and other polar interactions
5. Disulfide bridges
The image shows the two subunits with alpha units in red and beta units in yellow.
Recently, the nature of protein structure space has been widely discussed in the literature. The traditional discrete view of protein universe as a set of separate folds has been criticized in the light of growing evidence that almost any arrangement of secondary structures is possible and the whole protein space can be traversed through a path of similar structures. Here we argue that the discrete and continuous descriptions are not mutually exclusive, but complementary: the space is largely discrete in evolutionary sense, but continuous geometrically when purely structural similarities are quantified. Evolutionary connections are mainly confined to separate structural prototypes corresponding to folds as islands of structural stability, with few remaining traceable links between the islands. However, for a geometric similarity measure, it is usually possible to find a reasonable cutoff that yields paths connecting any two structures through intermediates
There has recently been much discussion on the origins of protein structure space. Researchers have been debating whether proteins are made up of discrete structure groups or a continuum. The traditional view of distinct structural folds has been questioned and many researchers are supporting the continuous view. The discrete view sees proteins as separate folds whereas continuous view supports the idea that any arrangement of secondary structures can be possible. Instead of debating on which of the two is correct, researchers have started to assert that both continuous and discrete views represent a duality in the sense that each view is necessary and present in protein structure space. Discrete and continuous views are actually complementary, protein structure space is discrete on an evolutionary level but continuous geometrically. Evolutionary connections are made by looking at certain folds as island of structural stability. To view protein structures for their geometric similarities, we see paths that connect any two structures through intermediates.
Discrete view:
The traditional discrete view was developed under the idea that there are many structural similarities present in protein structures. This idea was developed by utilizing X-ray crystallography to study the earliest protein structures, myoglobin and hemoglobin. Scientists have found that both myoglobin and hemoglobin have similar structures despite having different sequences. Other examples of structural similarities in protein structures include chymotrypsin and trypsin, several TIM beta/alpha barrels, Rossmann folds, and immunoglobin-like beta sandwiches. All of these structures are unique and recognizable..
The concept of ‘folds’ was developed to describe this discreteness in protein structure. Therefore, newly determined structures can be recognized as being one of these clear and recognizable structures or could be used to establish a ‘new’ fold so other protein structures could identify with it in the future. This idea was supported by the fact that most structures matched these commonly seen prototypes of protein structure.
The TIM-barrel fold is an example of a prototype. A great number of metabolic enzymes belong in the TIM-barrel fold prototype. Although most of these families did not show much similarity in terms of sequence, X-ray crystallographers found TIM-barrels in may enzyme families. These TIM-barrel families are known to have been created from gene duplications and build-up of metabolic pathways.
The fold discovered by Michael Rossmann is another example of a prominent structural prototype. This doubly wound fold is recognized by two right-handed βαβ units that are placed in a centrosymmetric form to make hydrogen bonds between the first strands of the units.
There is an evolutionary process that is occurring to evolve into a thermodynamically stable structure. Mutations that occur diverge from the evolutionary process by shying away from thermodynamic stability. Thus, these mutated proteins are eliminated by selection. The stabilizing selection that drives evolution makes it so movements between folds are uncommon. If we look at protein structure space in an evolutionary view, protein discreteness is characteristic of it.
Continuous view:
Many publications suggest that structure space is continuous. Similarity metrics like TM-score have allowed scientists to find a connection between any two structures through no more than seven steps.
Continuous space is not caused by evolution but by rules of hydrogen-bond formation and antiparallel and parallel arrangements of secondary structures. A continuous protein structure space also does not involve transitivity. The continuous view is more involved with geometry, which is also not transitive.
The term ‘fold’ has been questioned in a few publications on the account that categorizing structures into nonoverlapping folds can lead to missing important functional connections in different folds.
Grouping commonly seen structures with very distinct geometry can be very helpful for categorization and visualization. Using these groupings such as ‘TIM-barrel’. ‘Rossmann fold’, and ‘OB-fold’ that were discussed earlier could be helpful to determine structure and function. On the other hand, other types of ‘folds’ may be more ambiguous and only particular to a certain evolutionary group. Take proteins of different alpha-helical folds for instance. They are often characterized by progressive changes in the angles of helix packing in comparison to discrete topological differences. In cases of ambiguity such as these, the continuous view may be more appropriate. To determine protein function, it is necessary to study the lists of structure similarities rather than just fold assignments because placement and conformation of functional sites can be common even for protein structures with different folds. For example, TIM-barrels and Rossmann folds have discretely distinct geometries and their active sites have similar locations in between beta-strands and alpha helices.
‘Fold’ is a term that is essential to the SCOP classification (structural classification of proteins) and is a standard for making evolutionary connections between proteins. Proteins are said to have the same fold if they possess the same major secondary structural elements in the same mutual orientation and connectivity. This type of classification can cause problems because subjectivity is involved in choosing which secondary structure is major.
The definition of ‘fold’ is more of an empirical approximate ‘art’ because classification criteria for proteins are very loose and based on several things: structural data, and evolutionary and functional considerations.
Scientists are now using a revised term called ‘new fold’. Using this case, scientists are easy to miss meaningful connections between evolution and function with protein structure.
Scientists have questioned the immediate and practical value of protein structure space. They are fundamentally important, and especially necessary in structure prediction. The continuous viewpoint says that all structures are predictable if approximately 40% of overlapping structures are present. Another way these viewpoints can be applied is in predicting functional properties. Discrete and continuous views are both helpful for protein prediction. Both views are applicable in determining functionality, therefore one should not be seen as less important than the other, and neither should be criticized.
This idea of both continuous and discrete views playing a part in protein structure space suggests that there is a duality present in the nature of protein structure space. On an evolutionary standpoint, protein structure space is mostly discrete, and certain regions of stability correspond to certain protein folds. There are visible evolutionary connections between these regions. Geometrically speaking, the protein structure space is continuous. Any arrangement of secondary structures is possible, and almost any two structures can be connected by a path of intermediate and locally similar structures. An important note to remember about both discrete and continuous views is that homology is transitive, and homology is a property of discreteness; structure similarity is not. The continuous view is not transitive.
Model vs. Structure
[edit | edit source]Amidst research and study of proteins, there are confusions between 'structure' and 'models'. Often confused with structure, model refers to the arrangement of models based on the interaction of the molecules (which can include the side chains and bonds), homology between the molecules, or general reasoning. It is often part the data retrieved from experiments used to guide towards a model building. Models can be very simplistic or intricate. It can include arrows to represent the α or β structures, or arrows to show a path. They can also show the constrains of a measurement.
The structure of a protein refers to the spatial interaction of atoms with their covalent and non-covalentness. A structure is often obtained through experiments using x-ray crystallography, electron microscopy, NMR, etc. Electron microscopy and crystallography provide accuracy with the spatial resolution, while NMR experiments provide information on the atoms’ arrangements and their coordination. [1]
Chemical Reactions with Proteins
[edit | edit source]Proteins are made, modified, and identified in organic chemistry. Organic chemical reactions are necessary for proteins to exist and to function. The previous sections talk about reactions link together proteins. An important part of biochemistry and organic chemistry is to identify proteins and the sequence the amino acid are linked together. Identifying the sequences allows for comparison between other proteins. They are also valuable for making DNA and for encoding genes. The following techniques involve organic chemistry to help identify the sequence of proteins.
Edman Degradation
Edman degradation determines the amino acid at the N terminal. The reagent Phenyl isothiocyanate is used to cleave off the N terminal amino acid while leaving the rest of the chain intact. This is useful in identifying short peptides. However, it will take a long time to determine the sequence if the chain is long since it only identifies one amino acid at a time. Efficiency of this method decreases each time it is repeated. Therefore the best way to identify longer chains are to break them into smaller peptides for analysis.
Chemical cleavage
cyanogen bromide- CNBr cleaves chains at the carboxyl side of methionine
O-lodosobenzoate- cleaves carboxyl side of trypotophan
Hydroxylamine- cleaves asparagine- glycine bonds
2-nitro-5-thiocyanobenzoate- amino side of cysteine residues
Enzymatic cleavage
trypsin- cleaves carboxyl side of lysine and arginine
clostripain- cleaves carboxyl side of arginine residues
staphylococcal protease- carboxyl side of aspartate and glutamate
thrombin- cleaves carboxyl side of arginine
chymotrypsin- cleaves carboxyl side of tyrosine, tryptophan, phenylalanine, leucine, and methionine, mostly aromatic side chains
carboxypeptidase A- cleaves the amino side of C-terminal amino acid except arginine, lysine, or proline
Protein Degradation
[edit | edit source]Protein degradation is an important process that breaks down proteins into their smaller subunit amino acids. Degradation occurs in order to maintain and provide the body with a steady amount of amino acids. Misfolded or damaged proteins are broken down as they serve no purpose and must be destroyed. Dietary proteins are also degraded. Dietary proteins contain amino acids that cannot be synthesized in the body and therefore must be obtained through the foods we eat. These amino acids, known as essential amino acids, are a vital part in the resynthesis of new proteins.
Proteins are first digested in the stomach where the acidic conditions provide an optimal environment for the denaturing of the protein. The low pH enables proteolytic enzymes to unfold and degrade the protein. Pepsin is the main enzyme found in the stomach. Further break down of proteins occur in the lumen of the intestine. The pancreas provides the main source of the proteolytic enzymes that help in the break down of the protein. The ubiquitin-proteasome degradation of proteins is one pathway the body uses to regulate the supply of amino acids. A smaller protein called ubiquitin first tags the protein that needs to be degraded. This creates a marking on the protein that signals the proteasome to further digest it.
Once the protein is fully digested, the free amino acids counterparts are carried through the blood and delivered to necessary tissues to be absorbed and used for new protein synthesis. The free amino acids can also serve as energy in the process of cellular respiration. The amino group from the amino acid must first be removed so that the carbon skeleton can then be converted into a carbohydrate or fatty acid. The amino group is processed through the urea cycle where the nitrogen is disposed of.
Reference
[edit | edit source]Berg, Biochemistry, 6th Edition
http://faculty.clintoncc.suny.edu/faculty/michael.gregory/files/bio%20101/bio%20101%20lectures/biochemistry/biochemi.htm
[1] Fandrich, Marcus, Schmidt, Matthias, and Grigorieff, Nikoaus: Trends Biochem Sci. 2011 June; 36 (6) 338-345. “Recent Progress in understanding Alzheimer’s β-amyloid structures”