Structural Biochemistry/Proteins/X-ray Crystallography
X-ray crystallography
X-ray crystallography can reveal the detailed three-dimensional structures of thousands of proteins. The three components in an X-ray crystallographic analysis are a protein crystal, a source of x-rays, and a detector.
X-ray crystallography is used to investigate molecular structures through the growth of solid crystals of the molecules they study. Crystallographers aim high-powered X-rays at a tiny crystal containing trillions of identical molecules. The crystal scatters the X-rays onto an electronic detector. The electronic detector is the same type used to capture images in a digital camera. After each blast of X-rays, lasting from a few seconds to several hours, the researchers precisely rotate the crystal by entering its desired orientation into the computer that controls the X-ray apparatus. This enables the scientists to capture in three dimensions how the crystal scatters, or diffracts, X-rays. The intensity of each diffracted ray is fed into a computer, which uses a mathematical equation to calculate the position of every atom in the crystallized molecule. The result is a three-dimensional digital image of the molecule.
Crystallographers measure the distances between atoms in angstroms. The perfect “rulers” to measure angstrom distances are X-rays. The X-rays used by crystallographers are approximately 0.5 to 1.5 angstroms long, which are just the right size to measure the distance between atoms in a molecule. That is why X-rays are used.
Introduction
[edit | edit source]Protein X-ray crystallography is a technique used to obtain the three-dimensional structure of a particular protein by x-ray diffraction of its crystallized form. This three dimensional structure is crucial to determining a protein's functionality. Making crystals creates a lattice in which this technique aligns millions of proteins molecules together to make the data collection more sensitive. It's like getting a stack of papers, measuring the width with a ruler, and dividing that length with the number of pages to determine the width of one piece of paper. By this averaging technique, the noise level gets reduced and the signal to noise ratio increases.[2] The specificity of the protein's active sites and binding sites is completely dependent on the protein's precise conformation. X-ray crystallography can reveal the precise three-dimensional positions of most atoms in a protein molecule because x-rays and covalent bonds have similar wavelength, and therefore currently provides the best visualization of protein structure. It was the X-ray crystallography by Rosalind E.Franklin, that made it possible for J.D. Watson and F.H.C. Crick to figure out the double-helix structure of DNA.
Utilization
[edit | edit source]We use this procedure to grasp the cellular mechanism and the knowledge of the 3-D structure of enzymes and other macromolecules. It is critical that we can better understand how each chemical reaction that occurs in a cell needs a specific enzyme for it to happen. Two common techniques used for analysis of proteins structure are Nuclear Magnetic Resonance (NMR), and x-ray crystallography. X-ray crystallography can be used to analyze any different compounds up to a molecular weight of 106 (g/mol) for instance; where as NMR is restricted to biopolymers(polymers produced by a living organism such as starch, peptides, sugars) with a molecular weight no more than 30,000 (g/mol). It can also measure compounds that are very small because the appropriate size to measure the distance between atoms in a molecule is 0.5 to 1.5 angstroms. X-rays are used as the form of radiation because their wavelengths are on the same order of a covalent bond (~1 Å or 1 * 10−10m) and this is necessary to obtain a diffraction pattern that reveals information about the structure of the molecule. If the radiation had a wavelength much bigger or much smaller than the bond length of a covalent bond, the light would not diffract and no new knowledge of the structure would be obtained.
Techniques
[edit | edit source]The three components needed to complete an X-ray crystallography analysis are a protein crystal, a source of x-rays and a detector.
First Step
[edit | edit source]The process begins by crystallizing a protein of interest. Crystallization of protein causes all the protein atoms to be orientated in a fixed way with respect to one another while still maintaining their biologically active conformations - a requirement for X-ray diffraction. A protein must be precipitated out or extracted from a solution. The rule of thumb here is to get as pure a protein as possible to grow lots of crystals (this allows for the crystals to have charged properties, and surface charged distribution for better scattering results). 4 critical steps are taken to achieve protein crystallization, they are:
- Purify the protein. Determine the purity of the protein and if not pure (usually >99%), then must undergo further purification.
- Must precipitate protein. Usually done so by dissolving the protein in an appropriate solvent(water-buffer soln. w/ organic salt such as 2-methyl-2,4-pentanediol). If protein is insoluble in water-buffer or water-organic buffer then a detergent such as sodium lauryl sulfate must be added.
- The solution has to be brought to supersaturation(condensing the protein from the rest of the solvent forming condensation nuclei). This is done by adding a salt to the concentrated solution of the protein, reducing its solubility and allowing the protein to form a highly organized crystal (this process is referred to as salting out). Other methods include batch crystallization, liquid-liquid crystallization, vapor diffusion, and dialysis.
- Let the actual crystals grow. Since nuclei crystals are formed this will lead to obtaining actual crystal growth.
NOTES ON RECRYSTALLIZATION TECHNIQUE TO ACHIEVE A MORE PURE PROTEIN:
Recrystallization is an incredibly important technique used for the purification of substances. Understanding the solubility of the solid in a certain solvent is the key to recrystallization. One of the applications of this technique can be seen in pharmaceutics and in many other fields. For example, crystallographers use methods of nuclear magnetic resonance and x-ray diffraction to gain insight into different compounds. X-ray diffraction requires the formation of pure crystals in order to acquire accurate results. Crystallographers can gain insight into protein structure by using x-ray diffraction, but in order to be able to use x-rays to examine their crystals, they must first spend time forming pure protein crystals. It is very difficult to form protein crystals. It may even take years and incredibly specific conditions. Temperature, pH, and concentration have to be very specific to form larger crystals with a pure structure. Recrystallization in this process is vital to get rid of impurities in the crystal lattice. Scientists today use crystallography and recrystallization techniques to understand protein structure and help understand how a single abnormality in the protein's primary structure can cause diseases. All in all, purification techniques are vital in order to use x-ray diffraction to understand structure. In this experiment we explore the differences in micro and macro recrystallization. The techniques employed in recrystallization include finding a good solvent to work in, gravity filtration, slow cooling, and vacuum filtration. The key to a successful recrystallization is a good solvent. We need a solvent that will not dissolve the sample at cool temperatures but will dissolve it at high temperatures. This allows the precipitation of the solute after the solution is dissolved in warm temperatures. Since the solute is only soluble in the warm solute, upon cooling, a precipitate forms. Gravity Filtration is used to remove insoluble impurities remaining in the solution before recrystallization and it is used to filter out the charcoal used to remove the color impurities. Gravity filtration is effective, but we must avoid crystallization during this process as to avoid losing pure crystals in the filter paper. Slow cooling is also essential to ensure the purity and size of the crystals. When the solution is allowed to cool slowly, the dissolved impurities have time to interact with the solvent instead of remaining trapped in the crystal lattice. During fast cooling, impurities may remain trapped in the crystal lattice because crystallization occurs to quickly and impurities do not have time to return to the solvent. After the crystals are put in an ice bath to ensure maximum recrystallization, the solution is filtered using a vacuum filtration to extract the pure crystals from the solution with the impurities. After it is vacuumed, the pure crystals are collected and weighed. Micro recrystallization differs from macro recrystallization in the instruments and techniques used for filtration of the pure crystals. Micro recrystallization involves using a Craig tube and centrifugation instead of vacuum filtration. It is used for a recrystallization of less than 300mg of solid.
Second Step
[edit | edit source]For the next step, x-rays are generated and directed toward the crystallized protein. X-rays can be generated in four different ways,
- by bombarding a metal source with a beam of high-energy electrons,
- by exposing a substance to a primary beam of X-rays to create a secondary beam of X-ray fluorescence,
- from a radioactive decay process that generates X-rays (Gamma rays are indistinguishable from X-rays), and
- from a synchrotron (a cyclotron with an electric field at constant frequency) radiation source.
The first and last method utilize the phenomenon of bremsstrahlung, which states that an accelerating charge will give off radiation.
Then, the x-rays are shot at the protein crystal resulting in some of the x-rays going through the crystal and the rest being scattered in various directions. The scattering of x-rays is also known as "x-ray diffraction". Such scattering results from the interaction of electric and magnetic fields of the radiation with the electrons in the atoms of the crystal.
The patterns are a result of interference between the diffracted x-rays governed by Bragg's Law: , where is the distance between two regions of electron density, is the angle of diffraction, is the wavelength of the diffracted x-ray and is an integer. If the angle of reflection satisfies the following condition:
,
the diffracted x-rays will interfere constructively. Otherwise, destructive interference occurs.
Here is an example of constructive interference:
Here is an example of destructive interference:
File:Destructive Interference.jpg
Constructive interference indicates that the diffracted x-rays are in phase or lined up with each other, while destructive interference indicates that the x-rays are not exactly in phase with each other. The result is that the measured intensity of the x-rays increases and decreases as a function of angle and distance between the detector and the crystal.
The x-rays that have been scattered in various directions are then caught on x-ray film, which show a blackening of the emulsion in proportion to the intensity of the scattered x-rays hitting the film, or by a solid-state detector, like those found in digital cameras. The crystal is rotated so that the x-rays are able to hit the protein from all sides and angles. The pattern on the emulsion reveals much information about the structure of the protein in question. The three basic physical principles underlying this technique are:
- Atoms scatter x-rays. The amplitude of the diffracted x-ray is directly proportional to the number of electrons in the atom.
- Scattered waves recombine. The beams reinforce one another at the film if they are in phase or cancel one another out if they are out of phase. Every atom contributes to a scattered beam.
- Three-dimensional atomic arrangement determines how the beams recombine.
The intensities of the spots and their positions are thus the basic experimental data of the analysis.
Final Step
[edit | edit source]The final step involves creating an electron density map based on the measured intensities of the diffraction pattern on the film. A Fourier Transform can be applied to the intensities on the film to reconstruct the electron density distribution of the crystal. In this case, the Fourier Transform takes the spatial arrangement of the electron density and gives out the spatial frequency (how closely spaced the atoms are) in the form of the diffraction pattern on the x-ray film. An everyday example of the Fourier Transform is the music equalizer on a music player. Instead of displaying the actual music waveform, which is difficult to visualize, the equalizer displays the intensity of various bands of frequencies. Through the Fourier Transform, the electron density distribution is illustrated as a series of parallel shapes and lines stacked on top of each other (contour lines), like a terrain map. The mapping gives a three-dimensional representation of the electron densities observed through the x-ray crystallography. When interpreting the electron density map, resolution needs to be taken into account. A resolution of 5Å - 10Å can reveal the structure of polypeptide chains, 3Å - 4Å of groups of atoms, and 1Å - 1.5Å of individual atoms. The resolution is limited by the structure of the crystal and for proteins is about 2Å.
Type of X-ray Device
[edit | edit source]Protein molecules are very large, thus their crystals diffract x-ray beams much less than crystals from smaller molecules. Because larger molecules have fewer crystals, diffraction scattering and hence intensity emitted is very weak. Proteins contain carbon, nitrogen, and oxygen, and so are lighter elements(that is they have fewer electrons/atom); this is important since electrons are responsible for the diffraction and intensity, and therefore they scatter x-rays weaker than heavy elements. Knowing this, protein crystallographers use high intensity x-ray sources such as a rotating anode tube or a strong synchrotron x-ray source for analyzing the protein crystals.
- The number of electrons in an atom is proportional to the wave's amplitude. An example would be comparing a carbon atom and hydrogen atom, you would see that the carbon atom would scatter six times as strongly as the hydrogen atom.
- If in phase the waves combine with one another at the film but if the waves are out of phase then they cancel out one another at the film.
- The only thing that matters when looking at how scattered waves recombine is the atomic arrangement.
Energy of X-ray:
- where f is frequency and λ is wavelength. The SI unit of energy is the joule (J).
X-rays have higher energy than visible light due to its small wavelength.
Phase Problems
[edit | edit source]The interaction of X-rays with the electrons in a crystal gives rise to a diffraction pattern, which mathematically is the Fourier transform of the electron density distribution. The detectors used to measure the X-rays, however, can only measure the amplitude of the diffracted x-rays; the phase shifts, which are required to use the Fourier Transform and find the electron density distribution, are not measurable directly using this method. This is known in the physics community as the "Phase Problem". In simpler terms the phases cannot be found from the measured amplitudes of the X-rays. Other extrapolations must be made and additional experiments must be done in order to get an electron density map. Many times, the existing data on the compound's physical and chemical properties can help aid when there is a poor density map. Another method known as Patterson Synthesis is very useful to find out an initial estimate of phases and it is very useful for the initial stages to determine the structure of proteins when the phases are not known. The problem can be simplified by finding an atom, usually a heavy metal, using Patterson Synthesis and then using that atom's position to estimate the initial phases and calculate an initial electron density map that can further help in the modeling of the position of other atoms and improve the phase estimate even more. Another method is called Molecular Replacement; it locates the location of the protein structure in the cell. In addition to the molecular replacement method, the phase problem can also be solved by the isomorphous replacement method, the multiple wavelength anomalous diffraction method, the single-wavelength anomalous diffraction method, and direct methods.
Molecular Replacement
[edit | edit source]Phase problem can be solved by having an atomic model that can compute phases. A model can be obtained if the related protein structure is known. However, in order to build this atomic model, the orientation and position of the model in the new unit cell needs to be determined. This is when the technique, molecular replacement (or MR) comes in.
Molecular Replacement, also known as MR, is a method to solve phase problems in x-ray crystallography. MR locates the orientation and position of a protein structure with its unit cell, whose protein structure is homologous to the unknown protein structure that needs to be determined. The obtained phases can help generate electron density maps and help produce calculated intensities of the position of the protein structure model to the observed structures from the x-ray crystallography experiment.
MR method is also effective for solving macromolecular crystal structures. This method requires less time and effort for structural determination, since heavy atom derivatives and collecting data do not need to be prepared. The method is straight forward and model building is simplified because it needs no chain tracing.
This method consists of two steps:
- a rotational search to orient the homologous model in the unit cell or target
- a translational target where the new oriented model is positioned in the unit cell
Patterson-based (Molecular Replacement)
[edit | edit source]Patterson maps are interatomic vector maps that contain peaks for each related atom in the unit cell. If the Patterson maps were generated based on the data derived from the electron density maps, the two Patterson maps should be closely related to each other only if the model is correctly oriented and placed in the correct position. This will allow us to infer information about the location of the unknown protein structure with its cell. However, there is a problem with molecular replacement, it has six dimensions, three parameters to specify orientation and position. With the Patterson maps, it can be divided into subsets of the parameters to look at each part separately.
Rotation Function
[edit | edit source]Rotation function has intramolecular vectors that only depend on the molecule’s orientation and not its position because even when the molecule is translated in the unit cell, all of the atoms are shifted by the same amount but the vectors between the atoms are the same. The Patterson map for the unknown protein structure is compared with the homologous known protein structure in different orientations

Classic Rotation Function
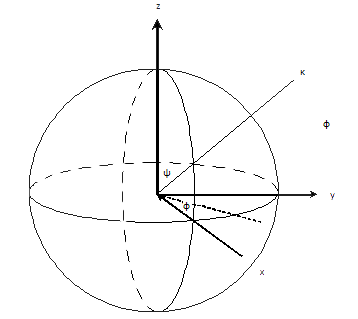
[edit | edit source]To find the orientation, determine the rotation axis and rotation angle about that axis. Two parameters will be needed to define an axis (a vector from the center of the sphere to a point on the sphere surface). The rotation axis starts off parallel to the z-axis and is rotated around the y-axis with angle ᶱ, then the object rotates around the z-axis with angle ᶲ, and finally it rotates around the rotation axis with angle ᵠ. These specify a point on the surface of a unit sphere.
Fast Rotation Function
[edit | edit source]The rotation function can be computed by comparing two Patterson maps or the peaks in those Pattersons. Rotation function can be computed much faster with Fourier transforms only if the Pattersons were expressed in terms of spherical harmonics.
Direct Rotation Function
[edit | edit source]In direct rotation function, the protein structure can be placed in the unit cell of the unknown structure and the Patterson for the oriented molecule is compared with the entire unknown structure Patterson.
Translation Function
[edit | edit source]Once the orientation of the known structure is known its model (electron density map) can be oriented to compute structure factors where a correlation function is used to determine the vector to translate the model on top of the homologous one within an asymmetric unit.
With the correct oriented and translated phasing models of the protein structure, it is accurate enough to derive the electron density maps from the derived phases. The electron density maps can be used to build and refine the model of the unknown structure.
Multiwavelength Anomalous Diffraction
[edit | edit source]X-Rays are generated in large machines called synchrotrons. Synchotrons accelerate electrons to nearly the speed of light and travel them through a large, hollow metal polygon-ring. At each corner, magnets bend the electron stream, causing the emission of energy in the form of electromagnetic radiation. Since the electrons are moving at the speed of light, they emit high energy X-rays.
The benefits of using synchrotrons is that researches do not have to grow multiple versions of every crystallized molecule, but instead only grow one type of crystal that contains selenium. They then have the ability to tune the wavelength to match the chemical properties of selenium. This technique is known as Multiwavelength Anomalous Diffraction. The crystals are then bombarded several times with wavelengths of different lengths, and eventually a diffraction pattern emerges which enables researchers to determine the location of the selenium atoms. This position can be used as a reference, or marker to determine the rest of the structure. The benefits of this allow researchers to collect their data much more quickly.
Isomorphous Replacement Method
[edit | edit source]This method compares the x-ray diffraction patterns between the original protein crystal and the same type of crystal with an addition of at least one atom with high atomic number. The method was used to determinate the structure of small molecules and eventually that of hemoglobin by Max Ferdinand Perutz (1914–2002). A perfect isomorphism is when the original crystal and its derivative have exactly the same conformation of protein, the position and orientation or the molecules, and the unit cell parameters. The only difference that the crystal and its derivative have in a perfect isomorphism is the intensity differences due to the addition of heavy atoms on the derivative. These differences can be identified manually or by an automatic Patterson search procedure, such as SIR 2002, SHELXD, nB, and ACORN, and such information is important as to determine the protein phase angles. However, perfect isomorphism hardly occurs because of the change in cell dimensions. For the protein with heavy atom, its tolerable change in cell dimension is dmin/4, for dmin is the resolution limit. Other factors, such as rotation, also contribute to nonisomorphism.
Procedures
[edit | edit source]- Prepare a few derivatives of the protein in crystalline structure. Then, measure the cell dimension to check for isomorphism.
- Collect x-ray intensity data of the original protein and its derivative.
- Apply the Patterson function to determine the coordinates of the heavy atom.
- Refine the heavy atom parameters and calculate the phase angle of the protein.
- Calculate the electron density of the protein.
The derivatives are made through two different methods. The preferred method is to soak the protein crystal in a solution that is composed identically to the mother liquor, but with a slight increase of precipitant concentration. Another method is co-crystallization, but it is not commonly used because the crystal will not grow or grow nonisomorphously. The soaking procedure depends on how wide the crystal pores are. The pores should be wide enough for the reagent to diffuse into the crystal and to reach the reactive sites on the surface of all protein molecules in the crystal.
Multiple Wavelength Anomalous Diffraction Method
[edit | edit source]Multiple Wavelength Anomalous Diffraction (abbreviated MAD) is a method utilized in X-ray crystallography that allows us to determine the structures of biological macromolecules, such as proteins and DNA, in order to solve the phase problem. Requirements for the structure include atoms that cause significant scattering from X-rays; notably sulfur or metal ions from metalloproteins. Since selenium can replace natural sulfur, it is more commonly used. The use of this technique greatly facilitates the crystallographer from using the Multiple Isomorphous Replacement (MIR) method as preparation of heavy compounds is superfluous.
this method is used to solve phase problems, when there is no available data regarding scattered diffraction besides amplitudes. Moreover, it is used when a heavy metal atom is already bound inside the protein or when the protein crystals are not isomorphous which is unsuitable for MIR method. The method has been mostly used for heavy metallo solution, these metallo enzyme normally comes from the 1st transition series and their neighbors. it is important to have a source for a powerful magnetic field to carry out this experiment, environment such as underground should be considered. A particle accelerator called a synchrotron is also required for the method.
Single-Wavelength Anomalous Diffraction Method
[edit | edit source]In comparison to multi-wavelength anomalous diffraction (MAD), single-wavelength anomalous diffraction (SAD) uses a single set of data from a single wavelength. The main beneficial difference between MAD and SAD is that the crystal spends less time in the x-ray beam with SAD, which reduces potential radiation damage to the molecule. Also, since SAD uses only one wavelength, it is more time-efficient than MAD.
The electron density maps derived from single-wavelength anomalous diffraction data do need to undergo modifications to resolve phase ambiguities. A common modification technique is solvent flattening, and when SAD is combined with solvent flattening, the electron density maps that result are of comparable quality to those that are derived from full MAD phasing. Solvent flattening involves adjusting the electron density of the interstitial regions between protein molecules occupied by the solvent. The solvent region is assumed to be relatively disordered and featureless compared to the protein. Smoothing the electron density in the solvent regions will enhance the electron density of the protein to an interpretable degree. This method is called ISAS, iterative single-wavelength anomalous scattering.
Direct Methods
[edit | edit source]The direct method can help recover the phases using the data it obtains. Direct Method estimates the initial and expanding phases using a triple relation. Triple (trio) relation is the relation of the intensity and phase of one reflection with two other intensities and phases. When using this method, the size of the protein structure matters since the phase probability distribution is inversely proportionate to the square root of the number of atoms. Direct method is the most useful technique to solve phase problems.
Interesting facts
[edit | edit source]Paul Peter Ewald and Max von Laue developed the idea to use crystals as a diffraction grating for X-rays in 1912. Von Laue proposed that compared to the larger wavelength of visible light, x-rays might have a wavelength close to the spacing of crystals' unit cells. He worked with Walter Friedrich and Paul Knipping to record the x-ray diffraction of a copper sulfate crystal onto a photographic plate. Von Laue developed a relation between the scattering angles and the size of the unit cell spacing and their orientation in the crystal, winning the Nobel Prize in Physics in 1914. As a result of von Laue's research, William Lawrence Bragg developed a law to connect a crystal's observed scattering and reflection from evenly-spaced planes in the crystal. This could be used to deduce atomic structure, and the significance of Bragg's Law to determining molecular structure was recognized immediately. In 1914, the first structure to be solved was that of table salt. Its electron distribution proved that not only covalent but also ionic compounds can form crystals.
In 1914, the structure of diamond was solved using x-rays, and it was shown that the length of the carbon-to-carbon single bond is 1.52 Angstroms.
X-ray crystallography was first used to determine protein structure in the late 1950s.
John Kendrew and Max Perutz while both at Cambridge used x-ray crystallography to discover the structure of hemoglobin & myoglobin(oxygen carrier in muscle) in 1945. They received the Nobel Prize in chemistry in 1962.
The first three-dimensional crystal structure of an enzyme determined via x-ray crystallography was a hen egg-white lysozyme. This was especially important as visual evidence of the transition-state theory because it was physical proof that the catalytic site was complementary to the transition-state geometry. Immediately following this first crystal structure, there was an upsurge in reports of the x-ray structures of many different enzymes.[3]
Examples
[edit | edit source]Many advances in drug discovery and medicine are due in large part by X-Ray Crystallography by identifying drug targets in many diseases that thrive today. In the late 80’s for example, scientists made a breakthrough in using X-Ray Crystallography to produce the structure of HIV Protease, an enzyme that was vital to the retrovirus’ life cycle. The enzyme cuts viral proteins strands that are main components of immature viral cells into separate, mature proteins that can continue on to form more mature and infectious viral particles. By looking closely at it structure, specifically its symmetry, researchers began making compounds that interacted with the active site of the enzyme, which is in the middle of its symmetric halves, to shut the enzyme down and prevent it from functioning properly. Amazingly, by the mid 90s, three HIV Protease inhibitor drugs were on the market, drastically reducing the death rate of the AIDS Virus.[4]
Not only is X-Ray Crystallography a useful tool for drug discovery, it is proven to be beneficial for making drugs better. For instance, current treatment for Parkinson’s disease involves inhibiting an enzyme called Monoamine Oxidase B [MAO B] that help recycle neurotransmitters by removing sometimes crucial molecular components that are left inactive. Although effective, such inhibitors cause undesirable side effects such as changes in heart rate, blood pressure, breathing, etc. However by determining the three dimensional structure of MAO B, along with seeing how some inhibitors attach to the enzyme, Dale Edmondson and his coworkers at Emory University have begun to contemplate methods of making new drugs that bind more specifically to the enzyme, in order to ultimately reduce the side effects.[5]
Additionally, X-ray crystallography has helped to explain how drugs work within the body, how they interact, what makes them work, and so on. A good example of such a case is the widely used drug aspirin. Aspirin has the ability to block the production of prostaglandins, messenger molecules that play various important roles in metabolism, by blocking the cyclooxygenase enzyme (COX) known to operate in the body's metabolic and immune systems. Scientists were able to study the COX enzyme and determine its structure via X-ray crystallography, and by doing so they got a clear picture of how the precise details of the enzyme's structure contribute to its overall molecular function. By determining the 3-dimensional structure of COX enzymes, we are able to understand how drugs like aspirin interact and block it.[6]
MIR Experiments
[edit | edit source]The MIR experiments, conducted by NASA, revealed that some proteins produce better quality crystals in a microgravity environment. DCAM, Ambient Diffusion-Controlled Protein Crystal Growth, was the procedure used in these experiments. DCAM uses a liquid to liquid diffusion method to grow protein crystals. These proteins were sent to the Mir Space Station by the Shuttle orbiter to crystallize. The protein crystals were later brought back to the ground for x-ray diffraction analysis. The results were promising. The largest crystals ever produced of certain proteins were produced by these experiments. These proteins include lysozyme, albumin and histone octamer. Bacteriorhodopsin, a membrane associated protein, produced crystals of improved size and quality. Furthermore, DCAM, performed in a microgravity environment, proved successful in providing unusually large protein crystals that could be analyzed for structure by neutron diffraction, a technology that cannot be utilized for smaller crystals grown on Earth. These promising results could produce numerous benefits for people on Earth. Protein crystals are needed to make therapeutic drugs. Scientists can develop new drugs to treat diseases by growing protein crystals in space that could not be grown in a large enough size on the Earth due to the limitations of gravitational forces.
Advantages with X-ray Crystallography
[edit | edit source]Some of the advantages of X-ray crystallography are that the technique itself can obtain an atomic resolution structure even if the atomic structure is in solution. This is because the structure in crystal form is the same if it were in solution.[Citation needed] Another advantageous aspect is that atomic structure contains a huge amount of data pertaining to the crystallized pure protein. The information one could receive from the structure of the protein can provide more information then finding its niche in the cellular environment.
Applications of X-ray Crystallography
[edit | edit source]The utilization of x-ray crystallography to determine protein structure has led to huge and significant breakthroughs in the area of structural biochemistry.
HIV
[edit | edit source]Although researchers have not found a cure for AIDS, structural biology has greatly enhanced their understanding of HIV and has played a key role in the development of drugs to treat this deadly disease. HIV was quickly recognized as a retrovirus, a type of virus that carries its genetic material not as DNA but as RNA. Long before HIV, researchers in labs all over the world studied retroviruses, some of which cause cancers in animals. These scientists traced out the life cycle of retroviruses and identified the key proteins the viruses use to infect cells. When HIV was identified as a retrovirus, these studies gave AIDS researchers an immediate jump-start making the previously discovered viral proteins the initial drug targets.
Scientists also determined the X-ray crystallographic structure of HIV protease, a viral enzyme critical in HIV’s life cycle, in 1989. Pharmaceutical scientists hoped that by blocking this enzyme, they could prevent the virus from spreading in the body. Scientists could finally see their target enzyme. By feeding the structural information into a computer modeling program, they could spin a model of the enzyme around, zoom in on specific atoms, analyze its chemical properties, and even strip away or alter parts of it. Most importantly, they could use the model structure as a reference to determine the types of molecules that might block the enzyme. Such structure-based drug design strategies have the potential to shave off years and millions of dollars from the traditional trial and error drug development process. The structure of HIV protease revealed a crucial fact, the enzyme is made up of two equal halves. For most such symmetrical molecules, both halves have an active site, that carries out the enzyme’s job but HIV protease has only one active site in the center of the molecule where the two halves meet. Pharmaceutical scientists can take advantage of this feature by blocking the single active site with a small molecule, they could shut down the whole enzyme and theoretically stop the virus’ spread in the body by using the enzyme’s structural shape as a guide.
Arthritis
[edit | edit source]Celebrex was initially designed to treat osteoarthritis and adult rheumatoid arthritis and became the first drug approved to treat a rare condition called FAP or familial adenomatous polyposis that leads to colon cancer. A fortunate discovery enabled scientists to design drugs that retain the anti-inflammatory properties of NSAIDs without the ulcer-causing side effects. By studying the drugs at the molecular level, researchers learned that NSAIDs block the action of two closely related enzymes called cyclooxygenases: COX-1 and COX-2. COX-2 is produced in response to injury or infection and activates molecules that trigger inflammation and an immune response. By blocking COX-2, NSAIDs reduce inflammation and pain caused by arthritis, headaches, and sprains. In contrast, COX-1 produces molecules called prostaglandins that protect the lining of the stomach from digestive acids so when NSAIDs block this function, they foster ulcers.
To create an effective painkiller that doesn’t cause ulcers, scientists realized they needed to develop new medicines that shut down COX-2 but not COX-1. Through structural biology, they could see exactly why Celebrex plugs up COX-2 but not COX-1. The three-dimensional structures of COX-2 and COX-1 are almost identical except for one amino acid change in the active site of COX-2 that creates an extra binding pocket. It is this extra pocket into which Celebrex binds. In addition to showing researchers in atom by atom detail how the drug binds to its target, the structures of the COX enzymes will continue to provide basic researcher with insight into how these molecules work in the body.
Problems with X-ray Crystallography
[edit | edit source]Some of the drawbacks of X-ray crystallography are that the sample needs to be in a solid form, the sample must be present in a large enough quantity to be studied, and the sample is often destroyed by the x ray radiation used to study it. This means that nothing in the gas or liquid state can be analyzed via x ray crystallography. Also, rare or hard to synthesize samples may be difficult to study, because there may not be enough of the sample for the radiation to provide a clear image. Thirdly, studying biological samples can be problematic because the radiation used to study the samples is most likely going to harm or destroy the living tissues.
One must also consider that x-ray crystallography takes a huge amount of time to complete upon one protein structure. The rough time estimates for each step of the process goes as follows: Cloning and purification of a protein structure to take up to 3-6 months with perfect execution and 99% purification. Crystallization can take up to 1-12 months pertaining to the physical properties of the protein which is completely based on the favorable enthalpy in which crystal formation is induced and the given solvent that the crystal is induced in. In addition data collection on the protein crystallized structure can take up to a month. Lastly, phasing the structure in solution can take about 3 months to complete. With this said, the process is not quick and with that comes financial issues when taking up a large time span and utilization of various laboratory equipment.
- To learn more about Protein X-ray Crystallography see Drenth, Jan: Principles of Protein X-Ray Crystallography 3rd ed.
References
[edit | edit source]- ↑ U.S. Department of Health and Human Services. The Structures of Life. July 2007.<http://www.nigms.nih.gov>.
- ↑ Viadiu, Hector. "Why do we need crystals?" UCSD Lecture. November 2011.
- ↑ Kraut, Joseph. "How Do Enzymes Work?", Science, vol.242, 28 October 1988, Pg.534
- ↑ National Institutes of Health, Structure Of Life, 2007, Pgs. 37-38, 40, 44.
- ↑ National Institutes of Health, “Medicine By Design, 2006, Pg.31.
- ↑ National Institutes of Health, “Medicine By Design, 2006, Pg.25-27.
- ↑ U.S. Department of Health and Human Services. The Structures of Life. July 2007.<http://www.nigms.nih.gov>.
- ↑ U.S. Department of Health and Human Services. The Structures of Life. July 2007.<http://www.nigms.nih.gov>.
Berg, Jeremy M., John L. Tymoczko, and Lubert Stryer. Biochemistry. 6th ed. New York: W. H. Freeman and, 2006. Print.








