Structural Biochemistry/Membrane Proteins/Ion Channels
Ion Channels
[edit | edit source]Ion Channels are porous transmembral proteins which help regulate the voltage gradient across the plasma membrane by allowing ions to diffuse through the pores down their electrochemical gradients. They all regulate the flow of ions across the plasma membrane with several protein subunits assembled in a circular arrangement with the narrowest area in the pore being only a few atoms across in diameter. They selectively allow ions to diffuse either by size or by charge. Other ion channels may act like a gate, allowing ions to pass through by electrical, temperature or other stimulus.
CNG Ion Channels
[edit | edit source]Cyclic-nucleotide channels, also known as CNG channels, are key in the sensory signaling of various types of cells, ranging from neurons to sperm cells. They come in two classes: classical CNG channels and CNGK- and mICNG – type channels. These two classes have helped activation mechanisms evolve overtime.
CNG channels produce voltage responses as they keep track of the intracellular concentration of cNMPs (cyclic nucleotides). Cyclic n[1]ucleotides channels are permeable to Ca2+ ions, and can carry a mixed cation current, therefore making them able to create a chemical symbol. Due to these characteristics, CNG channels play important roles in the signal pathways of retinal photoreceptors and olfactory receptor neurons. Photoreceptors in the retina respond to light and dark by opening or closing the CNG channels depending on cGMP (cyclic guanosine monophosphate) concentrations.
In the light, cGMP concentrations are low and photoreceptors go through a brief hyperpolarization and close the CNG channels. On the other hand, in the dark, cGMP concentration is high and CNG channels are open. When CNG channels are open, this allows a steady inward current. In contrast to photoreceptors, olfactory receptor neurons open CNG channels when they are being stimulate, whereas CNG channels are closed at rest.
CNG channels may also play a role in guiding nerve growth cones by utilizing molecules that attract and repulse in the nervous system. Another function CNG channels claim to have is control of chemotactic response of sea urchin sperm. Ca2+ oscillations are produced by a cGMP-signaling pathway, and those oscillations navigate the sperm.
Recent studies of the CNG channel family have found new members within bacteria and sea urchin sperm studies focusing on the function and structure of the new members show that binding a single molecule suffices to open CNG channels. There exist several other differences between the new CNG channels in comparison to the classical CNG channels.
Classical CNG channels are heterotetramers with subunits A and B. In mammals, four different A subunits and two different B units are known to exist in mammals.
There are two classes of CNG channels: low and high ligand sensitivity. Differences in ligand sensitivity are organized by the different interactions between ligand and CNBD. Another way ligand selectivity can be determined is by a specific Asp residue in the CNGA1 channel. Side chains that are negatively cGMP selectivity and reduces the cAMP gating efficacy.
New CNG (cyclic nucleotide-gated ion channels) have been found in sea urchin sperm and bacteria that are uncooperative because they only need a ligand molecule to bind in order to open the gate.

For photoreceptors in the eye, there is a high concentration of cGMP in dark; the light creates hydrolysis at phosphodiesterase. For olfactory receptor neurons, the odorants start the synthesis of cAMP.

CNG polypeptides contain transmembrane domains named S1-S6. S4 which detects voltage in Ca, Na, and K channels has a chain of charge Lys or Arg residues spread out between hydrophobic amino acids. This is unlike the classic CNG channel which does not rely as heavily on voltage. Glutamic acid is a key component of CNGs: in the hairpin turn portion containing pores, the Glu is carried as residue; at the tetramer, a ring of Glu provides an area for organization and movement of Ca+2 ions.
At the C-terminus, the cyclic nucleotide-binding domain or CNBD is attached to S6 by about 80 amino acids which convey the binding of the ligand with the gate of the channel. It is thought that this link supports tetramerization.
At the A and B parts of the CNG, calmodulin binding (CaM) with Ca+2 dictates the channel’s activity. Four of these CaM binds have been found in olfactory neurons, but only those at B and A4 seem to matter due to their N and C-terminus binding. The binding between CaM and Ca+2 has been found to lower sensitivity to ligands, offering a smoother transition to the resting state.
The mlCNG channel from the bacteria Mesorhizobium loti is like classic CNG channels in the sense that it creates tetramers and is made from six transmembrane segments with a C-terminus CNBD. It differs because it contains the GYGD sequence of K channels (Gly-Tyr-Gly-Asp). Also, instead of being about 80 amino acids, the C-link is about 20 amino acids which cause it to not support tetramerization.
CNGK structures are CNG channels which has the sequence of the K channel in its pore region. CNGK’s have a large polypeptide which holds four areas with a similar sequence or ‘repeats’.
Scientists are trying to see how it would be possible to open all four gates at once. They use double electron-electron resonance (DEER) to give the structure and point out where residues and conformation movements are.
CNG channels help us understand the relationship between ligand binding and gates. [2]
Potassium Channel
[edit | edit source]Potassium channels are found basically in most organisms. Potassium channels must be ancient because many channels had originated from them. Generally, there are two types of potassium ion, the six-transmembrane helix voltage gated and the two-transmembrane helix inward rectifier. Usually, potassium channels have the amino acid sequence of TMxTVGYG. In addition, the six-transmembrane helix voltage gated type typically has the unique pattern of lysine and arginine show up at every third or fourth place in the sequence. Scientists had studied potassium in a bacterial protein called KcsA and found that its structure is very similar to that of eukaryotic. Thus, they used KcsA as a model to study about K+ channel.
Potassium channel in a shape of a pore locates in the plasma membrane but not in a random manner. It carries an important role for it contributes in many different processes in the body such as secreting of hormones, regulating vascular system, etc. Potassium channel is also important in plant for guard cells uses K+ channels to control osmotic flow. Potassium channel has two components to it: the filter and the gate. The filter is responsible for choosing the ion, which in this case is potassium, and the gate opens and closes depending on the outside environment such as the effect of voltage or signaled molecules. The structure of the filter is similar to that of hemoglobin in the way that it is a tetramer with four repeating subunits that surround the pore. Also, this structure is known to be asymmetric. Moreover, the subunits are hydrophilic. The flow of potassium ion through the channel happens very quickly. Although the name is potassium channel, it also allows sodium ion to pass through. However, the ratio of potassium to sodium is 10,000 to 1. Another reason why sodium ion wants to get through the channel is because the ions are competing against each other.
Since potassium channel relates to nerve signaling and if the channel does not function normally, this can result in serious damage. What makes potassium channel only permeable to K+ ion is the imitation of oxygen across the channel. Potassium is known for having water around it. The formation of oxygen on the inside wall of the channel facilitates the flow of potassium ion through the channel. Even though much is known about potassium channel, the function of K+ channel in prokaryotic cell is still a mystery.
M2 Proton Channel
[edit | edit source]M2 proton channel resides in the envelope of the virus influenza A. The structure of the M2 proton channel is a homotetramer interconnected by disulfide bonds. is similar to that of potassium channel in which is it also a tetramer, and it consists of four similar subunits. The subunits in this case is the M2 itself. Moreover, the subunits take the shape of helices and are bonded together by disulfide bonds. M2 proton channel functions best when the pH level is low. The M2 protein is composed of three components: the 24 amino acids of the N terminal which face the outside of the cell, the 22 amino acids which are embedded on the membrane and are hydrophobic, and the 52 amino acids of the C terminal which face the cytoplasm. [3]
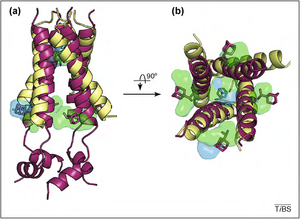
The 22 amino acid chain forms a trans membrane alpha helix. Since the amino acid Val27 is close to the N terminal the M2 channel is very selective in term of protons. As a result, protons that go through the channel can only pass through in the form of water molecule, and it is not easy for molecules such as hydrated sodium to pass through.
Activation of the proton channel occurs from low pH. The main function of the M2 channel is to allow hydrogen ions from the endosome to enter the virion and lower the pH within the virus. This causes disassociation of the matrix protein surrounding the virus’s genome, which is crucial in exposing the virus’s contents to the host cell’s cytoplasm.[4] The two medicines that were designed to stop the function of M2 channel are amantadine and rimantadine. These two medicines prevent the attack of the virus on the host cell. Nevertheless, because M2 is prone to be affected by mutations, the use of these medications becomes helpless. For instance, if one amino acid in the membrane is changed, the drugs will not be able to block the virus anymore. Two structures of M2 have been discovered: one is mutated and locates in the membrane, the second structure is similar to the first except it is longer, and it has the C-terminal component. Due to the virus’s susceptibility to mutations, substitutions along the trans membrane alpha helix have led to the virus’s immunity to inhibitors. In particular, the Ser31Asn substitution renders the protein highly resistant to amantadine inhibition. This most recently occurred during the 2009 ‘swine flu’ pandemic where 100% of samples tested during the season were resistance to amantadine and its derivative rimantidine. [5]
History
[edit | edit source]Scientists began to understand the structure and function of ion channels beginning in the early 1950s when British biophysicists Alan Hodgkin and Andrew Huxley analyzed the properties of currents by ion channels in their 1951 research on action potentials which won them the Nobel Prize. The existence of ion channels were indirectly confirmed by Bernard Katz and Ricardo Miledi in 1971 and later were directly proved by Erwin Neher and Bert Sakmann by using the patch clamp technique. In 2003, the Nobel Prize in Chemistry was awarded to Roderick MacKinnon for x-ray crystallographic studies on ion channel structure and to Peter Agre for similar work on aquaporins.
Biological Importance
[edit | edit source]Ion channels are prominent features of the nervous system which allow for conduction between the synapses. In addition, ion channels facilitate rapid changes in cells and are therefore useful as a target for new drugs. However, the importance of ion channels for nerve cell function result in evolution of toxins secreted by animals and plants which target the central nervous system and result in paralysis. Some examples include tetrodotoxin used by pufferfish for defense and novocaine which block sodium channels. Others include dendrotoxin which is produced by the snake, the Black Mamba, which blocks potassium ion channels. In addition, genetic mutations in DNA which encode for the ion channel protein would cause the same type of reactions.
Notes
[edit | edit source]- ↑ http://www.ncbi.nlm.nih.gov/pubmed/20729090
- ↑ http://www.ncbi.nlm.nih.gov/pubmed/20729090
- ↑ Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF (2008). "Structural basis for the function and inhibition of an influenza virus proton channel" (PDF). Nature. 451 (7178): 596–9. doi:10.1038/nature06528. PMID 18235504.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Helenius, A. (1992) Unpacking the incoming influenza-virus. Cell 69,577–578
- ↑ Crosby, Niall J; Deane, Katherine; Clarke, Carl E (2003). Clarke, Carl E (ed.). "Amantadine in Parkinson's disease". Cochrane Database of Systematic Reviews. doi:10.1002/14651858.CD003468.