Proteomics/Protein Identification - Mass Spectrometry/Data Analysis/ Interpretation
This Section:
Data Analysis
[edit | edit source]Mass Spectrum
[edit | edit source]A mass spectrum is a plot of an intensity vs. mass-to-charge ratio of a separated chemical collection. The mass spectrum of a given sample is the distribution pattern of the components of that collection, whether atoms or molecules, based their mass-charge ratio.
The X-axis of the plot is the mass-charge ratio also seen as (m/z) which is the quantity obtained by dividing the mass number of an ion by its charge number. For mass analyzers such as Time of Flight, the direct X-axis measurement is the time series of the ions measured by the detector. For such cases, the spectra must be calibrated with known standards in order to transform the X-axis from a time series into a m/z ratio. The values for the standards are used to generate the parameters for the equations relating the time of flight to m/z ratio. After these parameters are determined, the m/z ratios for the unknown sample can be calculated from their time of flight. With the Fourier Transform Ion Cyclotron Resonance mass spectrometer, the frequency measurements gathered by the detector plates undergo Fast Fourier Transformation before they are mass calibrated.

The Y-axis of a mass spectrum represents the signal intensity of the ions, and has arbitrary units. In most forms of mass spectrometry, the signal intensity of an ion current does not represent relative abundance accurately, but somewhat correlates loosely with it. Signal intensity is dependent on certain factors, the nature of the molecules being analyzed, how they ionize, the buffers' interaction, and the sample interaction.
Method of Interpretation
[edit | edit source]Samples can sometimes be highly difficult to analyze due to the amount of restrictions as well as variables that play roles in the output. Many factors can play roles in how a mass spectrum is interpreted, these factors may include: even electron vs. odd electron species, positive vs. negative ion mode, intact protein vs. fragmented peptide ions, etc.
Basis of resolution as well as peak height is reliant on the amount of sample being used and the amount of separation done prior to the mass spectroscopy. Based on certain heights and areas or peaks, structures can be determined.
Not all mass spectra can be interpreted similarly, due to the varying nature of the mass analyzers and ionization methods available. For example, some mass spectrometers break the analyte molecules into fragments; others observe the intact molecular masses with little fragmentation. A mass spectrum can represent many different types of information based on the type of mass spectrometer and the specific experimental conditions applied; however, all plots of intensity vs. mass-to-charge are referred to as mass spectra.
Normalization Techniques
[edit | edit source]
The process of removing statistical error from data created from repeated measurements is called normalization. In MS normalization techniques are used to remove systematic biases from peptide samples. These biases can arise from various sources including protein degradation, measurement errors, and variation in loading samples. The common normalization techniques used on MS data require that the data is transformed from the linear to the log scale. Doing so allows the values to conform to the normal distribution and reduces the likelihood of masking more relevant proteins with less relevant ones.
Annotation
[edit | edit source]As described in the section above, there are a number of factors that are vital to meaningful interpretation of the data from a mass spectrometry experiment. Without this metadata about the variables of the experiment, it is difficult to use a mass spectrum to generate an assessment. Combine this with the variance among types of mass spectrometers, and the utility of a reporting standard emerges.
In the vein of Minimum Information About A Microarray Experiment(MIAME), the HUPO Proteomics Standards Initiative developed MIAPE MS, a Minimum Information About a Proteomics Experiment in Mass Spectrometry. The standard requires metadata to be recorded about general information such as the machine manufacturer and model, variables of the ion source for ESI and MALDI, mass analyzers and detectors involved, and post-processing involved in peak list generation and annotation. In addition to the mass spectrometer operation, post-processing methods that are often used in other fields of signal processing are frequently applied to mass spectra in order to generate a more useful spectrum.
Further work has been done to regulate the expression of Mass Spectroscopy data via the establishment of a Controlled Vocabulary (CV) [1]. Developed by HUPO under the Proteomics Standards Initiative, the CV provides an ontology, comprised of applicable terms and transitions, which will better control the representation of Mass Sepctrometry data by regulating the vocabulary used in their description.
Accurate Mass Time Tags
[edit | edit source]
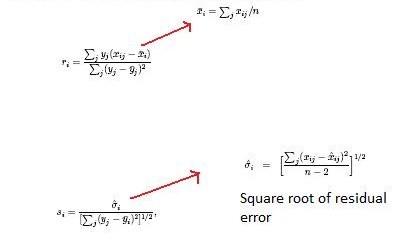
The accurate time tag approach, or AMT, takes advantage of the highly accurate readings from FTICR-MS and nanoLC. Data from these techniques can be used to form unique tags based on their molecular mass and retention times for a specific peptide. This technique assumes that there is enough separation power in two dimensions that it is very unlikely that two will occur with the same mass and separation time. Thus is it unlikely that a new species will be detected upon further analysis of the same sample with the same mass and retention time as any other species.
Decision Tree Ensembles
[edit | edit source]
Decision tree is one of the machine learning methods widely used to analyze proteomics data. Generated from a given dataset, a single decision tree reports a classification result by each of its terminal leaves (classifiers). Even though there exist many algorithms such as C4.5 that can be used to generate a well-modeled single decision tree, it is still possible that its prediction is biased, thus adversely affecting its accuracy.
To overcome this problem, more than one decision trees are used to analyze the data. It is based on the concept of forming a panel of experts who will then vote to decide the final outcome. The panel of experts is analogous to the ensemble of decision trees, which provide a pool of classifiers. Similar to voting, the classifier that has a majority becomes the true classification result for the data. As reported by Ge et al.,[1] the decision tree ensemble is more accurate than a single decision tree.
The diagram shown summarizes the process of generating decision tree ensemble and classification of data using Bagging (bootstrap aggregating) algorithm. To explain briefly, Bagging algorithm begins with randomly sampling (with replacement) the data from the original dataset to form a training set. A multiple of training sets are usually generated. Note that, since the replacement is allowed, the data in the training set can be duplicated. Each of training sets then generates a decision tree. For a given test data, each decision tree predicts an outcome, represented by a classifier. The ensemble of decision trees forms a panel of experts whose votes determine the final classification result from this group of classifiers.
Significance Analysis of Microarray Applied to MS data
[edit | edit source]While the Significance Analysis of Microarray (SAM) method was intended for the identification of differential gene expression, it has recently been utilized for identifying differential protein expression with data obtained from liquid chromatography mass spectrometry. Proteomic data retrieved from nanoLC/LTQ/FTMS and analyzed via SAM's algorithm has revealed reliable data not only relating to differential protein expression, but in determining false discovery rates with a level of accuracy previously unmatched by standard proteomic analysis tools. The superiority of this method over other methods of analysis such as identifying genes as significantly changed if an R-fold change is observed and significant changes in expression based on if an R-fold change was seen consistently between paired samples were discussed. Additionally, information regarding obtaining and using the SAM program is available (Section 1.11.4).
Mass Spectra Data Formats
[edit | edit source]mzData
[edit | edit source]HUPO PSI developed this XML format meant to unify the existing formats into a standard format for reporting peak list information called mzData. Essentially mzData provided a standard meas to represent peak data from a Mass spectroscopy experiment, and establish the fundamental parameters necessary for proper interpretation of the spectra, such as negative or positive ion spectra, and other basal information.
mzXML
[edit | edit source]Developed by the Seattle Proteome Center at the Institute for Systems Biology this data format is similar in its minimal requirements for the presentation of MS data, but with several key differences which make it the preferred file format for MS data [2]. Numerous converters and tanslaters exist which allow for the ubiquitous data transfer from a variety of the major Mass Spectrometers including Waters, Thermo, Bruker, MDS, Agilent, and ABI. Based from the same data structure as mzXML, prepXML (also produced by the Seattle Proteome Center), is a data format that has arisen to model protein sequencing data generated from MS-MS experimentation. Additional data formats for only protein and peptide data, such as protXML (Seattle Proteome Center), exist for purpose driven representation of protein MS experiments.
mzML
[edit | edit source]Resultant of the HUPO-Proteomics Standards Initiative's continuing progress towards standardization of proteomics data, specifically Mass Spectroscopy data, led to the development of a new unifying Mass Spectra data format called mzML. Simply put this data format attempts to combine the existing standards for both Mass Spectrometry experimental design as well as the resulting peak information.
References
[edit | edit source]- ASMS - What are the characteristics of a mass spectrum?, http://www.asms.org/whatisms/p5.html
- Agilent Technologies MS Explained - Spectrum
- McLafferty, F. W. and Turecek, F., Interpretation of Mass Spectra, University Science Books; 4th edition (May, 1993) ISBN 0935702253
- Zimmer JSD, Monroe ME, Qian WJ, Smith RD, Advances in Proteomics Data Analysis and Display Using an Accurate Mass Time Tag Approach. Mass Spectrom Rev. 25(3):450-482 (2006)
Articles Summarized
[edit | edit source]Normalization Approaches for Removing Systematic Biases Associated with Mass Spectrometry and Label-Free Proteomics
[edit | edit source]Stephen J. Callister et al. J Proteome Res. Volume: 5(2) 227-286 (2006)[2]
Main Focus
The focus is to attempt to find a technique for statistically normalizing MS data so that it can be meaningfully analyzed.
Summary
When looking at Mass Spectrometry (MS) data, the results can never be replicated, even when running replicate samples. Thus, there needs to be a way to make the results comparable. Since the difference in results can be related to bias and noise in addition to biological change, causing extraneous variability, normalization techniques are necessary. Four different normalization techniques commonly used in Microarray Analysis (central tendency, linear regression, locally weighted regression, and quantile techniques) were tested on three different sample sets (standard proteins, an early-log growth phase D. radiodurans sample and a stationary growth phase sample, and a striata brain sample of a control mouse and a striata brain sample of a methamphetamine-stressed mouse), which showed different levels of protein complexity.
Although all of the techniques did reduce systematic bias at least somewhat, because there was no definitive trend among the results obtained by the different techniques, none of these can yet be used for normalizing MS data; however, this study provides guidance for developing proper normalization techniques.
New Terms
- Bias
- variation arising from systematic errors in the experiment, sample preparation, or instrumentation (http://www.onesmartclick.com/exams/statistics-bias.html)
- Noise
- variation arising from random errors in the experiment, sample preparation, or instrumentation
- Central tendency
- this technique centers the abundance of a peptide around the mean or other constant to adjust for independent systematic bias (http://cnx.org/content/m10942/latest/)
- Local Regression
- technique that assumes that the systematic bias is not linearly dependent on the abundance of the peptides (http://www.biostat.jhsph.edu/~ririzarr/Teaching/754/section-03.pdf)
- Quantile
- nonparametric statistical approach which was originally designed for use with multiple high density arrays (http://mathworld.wolfram.com/Quantile.html)
Course Relevance
Mass spectrometry (MS) is one of the main tools of Proteomics, as it provides a way to obtain measurements of the abundance of individual proteins in the sample. Since the goal of Proteomics is to obtain measurements of the abundance of the different expressed proteins in different conditions, MS is a powerful tool.
Advances in Proteomics Data Analysis and Display Using an Accurate Mass Time Tag Approach
[edit | edit source]Zimmer JSD, Monroe ME, Qian WJ, Smith RD Mass Spectrom Rev. 25(3):450-482 (2006)
Main Focus
Recent advances in proteomic technologies have provided tools which provide both high efficiency and high-throughput proteomic analysis. These tools, specifically nanoLC-FTICR-MS and the necessary data processing and management tools, are the focus of this article.
Summary
While the field of proteomics is relatively new techniques have existed for 30 years which can now be applied to these new proteomic problems. These techniques, such as Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS), provides both high sensitivity and high mass measurement accuracy (MMA) necessary for identification of species as well as a wide dynamic range. FTICR-MS is also well suited for both “top-down” and “bottom-up” proteomics in so much as it can determine protein/peptide identities from parent masses as well as based upon fragmented patterns, which can deal with very complex mixtures of peptides. In addition to the difficulties in the production of high throughput techniques, problems also exist in the data management of the collected data. Results from a single experiment using high performance FTICR-MS can typically produce a 10GB raw data file, much too large for storage of any significant volume. Using techniques which take advantage of low statistical probabilities existing that new species detected in a sample will be detected in additional analyses of the same system. Using clustering in n-dimensional space using the Euclidean distance sets of unique mass classes can be produced reducing redundancy. Clustering is necessary due to the unreliable reproduction of data from LC-MS. Typically shifts in elution time occur and differ between multiple runs of the same peptides, where one would expect the same result. These shifts are the result of flow rate, temperature, differences in the packing of the columns as well as contamination. These same problems can also exacerbate the process of normalization of the data sets. Sample preparation is a key step in the proteomic analysis and can be quite difficult. Protein concentrations can vary greatly with only slight environmental changes. Many such techniques have been developed, such as solid phase isotope-coded affinity tags (SPICAT) for quantitative analysis, post-digestion trypsin-catalyzed 16O/18O labeling which has its advantages in that all types of samples can be labeled in the manner, quantitative cysteinyl-peptide enrichment technology (QCET), for use in high throughput experiments involving mammalian cells. Each of these techniques has their own limitations and steps are being taken to overcome these limitations. Additionally methods have been developed to expand the dynamic range and minimize noise created by abundant species of a sample. Dynamic range enhancement applied to mass spectrometry (DREAMS) aids in the detection of species which are both biologically significant and have low relative abundance on the fly. Targeted, or data-directed, LC-MS/MS can focus on a subset of proteins that have significant changes in abundance between two samples, and multiplexed MS/MS provides a way to increase the rate and sensitivity of proteomic measurements. These techniques have a wide range of applications already being thoroughly researched as well as adapt current techniques to better understand the biological processes which require the presence of these proteins or changes in their relative abundance.
New Terms
- Euclidean distance
- The straight line distance between two points. In a plane with p1 at (x1, y1) and p2 at (x2, y2), it is √((x1 - x2)² + (y1 - y2)²). ( http://www.itl.nist.gov/div897/sqg/dads/HTML/euclidndstnc.html )
- Normalization
- The process of identifying and removing the systematic effects. ( http://www.absoluteastronomy.com/topics/Normalization_(statistics) )
- Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS)
- A type of mass analyzer (or mass spectrometer) for determining the mass-to-charge ratio (m/z) of ions based on the cyclotron frequency of the ions in a fixed magnetic field. (http://www.ncbi.nlm.nih.gov/pubmed/9768511?dopt=Abstract)
- Elution
- the process of extracting one material from another by washing with a solvent to remove adsorbed material from an adsorbent. (http://wordnetweb.princeton.edu/perl/webwn?s=elution)
- Clustering
- The goal of data clustering, or unsupervised learning, is to discover "natural" groupings in a set of patterns, points, or objects, without prior knowledge of any class labels. (http://dataclustering.cse.msu.edu/)
Course Relevance
- Techniques which are both highly efficient and high-throughput are necessary for analysis of large data sets, such as the proteome. Without these tools analysis of the proteome would be too slow.
Classification of Premalignant Pancreatic Cancer Mass-Spectrometry Data Using Decision Tree Ensembles
[edit | edit source]Ge G, Wong GW. BMC Bioinformatics 9:275 (2008)
Main Focus
To compare the performance of several machine learning algorithms based on decision tree, Ge et al. conducted a series of statistical analyses on mass spectral data, obtained from a premalignant pancreatic cancer research. It was found that the classifier ensemble techniques outperformed their single algorithm counterparts in terms of the consistency in identifying the cancer biomarkers and the accuracy in the classification of the data.
Summary

In this article, Ge et al. reported that the performance in identifying biomarkers for premalignant pancreatic cancer could be enhanced by using the decision tree ensemble techniques instead of a single algorithm counterpart. These techniques had proven more likely to accurately distinguish disease class from normal class as indicated by a larger area under the Receiver Operating Characteristic curve. Moreover, they achieved comparatively lower root mean squared errors.
According to their method, the peptide mass-spectrometry data were processed first to improve data integrity and reduce variations among data due to the differences in sample loading conditions. The preprocessing steps involved baseline adjustment using group median, smoothing to remove noise using a Gaussian kernel, and normalization to make all the data comparable. After that, the data were randomly sampled such that 90% formed a training set and the remaining 10% formed a test set.
The training set was used in feature selection. In the study, the authors considered three different feature selection methods. The first method was a two-sample homoscedastic t test, which was used under the assumption that all the features from either normal or disease class had normal distribution. Unlike the first method, the second method based on Wilcoxon rank test considered that the features had no distribution. The last feature selection method was a genetic algorithm.
The test set was used to generate a single decision tree including the decision tree ensembles. The ensemble methods being studied were Random Forest, Random Tree, Bagging, Logitboost, Stacking, Adaboost, and Multiboost. Their performances were measured in terms of accuracy and error in the classification of the features, selected by each selection method. Then, they were compared against the performance of a single decision tree generated by C4.5 algorithm. The process repeated ten times to validate the resulting performance consistency.
According to the results reported, the decision tree ensembles achieved higher accuracy up to 70% regardless of the feature selection methods used. In terms of biomarker identification, both the t test and the Wilcoxon rank test had similarly impressive performance by consistently selecting the same biomarker-suspect features. Unlike the first two methods, the performance of the genetic algorithm was considerably poor. Ge et al. also noted that 70% accuracy was still lower than expected. This could be as a result from a naturally low concentration of the biomarkers at the premalignant stage of the cancer. In addition, it was also possible that one dataset might not be suitable for all algorithms, thus underestimating the accuracy.
New Terms
- Biomarker
- Bio-molecules that can be used to distinguish an abnormal from normal process, or a disease from condition. They can also be used as an indicator for a particular process such as drug interaction. These bio-molecules are usually found in blood, other body fluids, or tissues ( http://www.cancer.gov/dictionary/?searchTxt=biomarker )
- Receiver Operating Characteristic curve
- A graphical plot of the sensitivity vs. 1-specificity for a binary classifier system for different thresholds ( http://en.wikipedia.org/wiki/Receiver_operating_characteristic )
- Gaussian kernel smoothing
- A process of averaging the data points by applying a Gaussian function. Basically, the Gaussian function is used to generate a set of normalized weighting coefficients for the data points whose weighted sum generates a new value. This new value replaces the old one at the center of Gaussian curve. ( http://imaging.mrc-cbu.cam.ac.uk/imaging/PrinciplesSmoothing )
- Homoscedastic
- A sequence or a vector of random variables that have the same finite variance for all random variables ( http://en.wikipedia.org/wiki/Homoscedasticity )
- C4.5 algorithm
- An algorithm used to generate a decision tree from a set of training data (a set of classified samples) ( http://en.wikipedia.org/wiki/C4.5_algorithm )
Course Relevance
- The feature selection methods and decision tree ensembles introduced in this article contribute an interesting methodology for data analysis to the mass spectrometry topic in proteomics class.
Significance Analysis of Microarray for Relative Quantitation of LC/MS Data in Proteomics
[edit | edit source]Main Focus
The Significance Analysis of Microarray (SAM) analysis method which is normally used in the analysis of DNA microarrays to identify differential gene expression may also be used to identify differential protein expression. This analysis method can identify false positive results more accuretly than the conventional test generally used for this purpose.
Summary
The Significance Analysis of Microarray (SAM) analysis method was developed by researchers at the Stanford University to analyze genes in microarrays in order to identify genes that are differentially expressed (statistically) and obtain accurate false discovery rate statistics. In "Significance Analysis of Microarray for Relative Quantitation of LC/MS Data in Proteomics" (Li et al.) the SAM method is applied to proteomics data retrieved from nanoLC/LTQ/FTMS in order to determine differential expression for the proteins in a biological sample. SAM was also used to determine and estimate of rates of false discovery as well as miss rates. SAM results were them compared to results obtained from more tradition proteomics analysis tools such as the conventions t-test and by fold change. The biological system that was used to test the analysis techniques involved growing mycobacterium smegmatis cultures at pH5 and pH7, and looking for differences in protein expression under these two conditions. They compared protein abundance and focused on changes in protein expression and rates for false positives. The analysis appears to reveal that the SAM method can "zero in" on false positives more accurately than the t-test, making it the more accurate test, and allowing protein changes to be identified with a 5% false positive rate. The chart below contains data showing that a larger number of differentially expressed proteins were found using SAM, while maintaining a low false positive rate.
New Terms
- nanoLC/LTQ-FTMS
- This stands for nanoliquid chromatography/linear ion trap-fourier transform mass spectrometry. This type of Mass spectrometry analysis combines the high quality and reproducible data obtain from nanoLC with the power of a linear ion trap quadrapole, and accuracy of Fourier transform analysis. This is a very powerful proteomics tool. (https://products.appliedbiosystems.com/ab/en/US/adirect/ab?cmd=catNavigate2&catID=601452&tab=DetailInfo)
- t- test
- A statistical test involving means of normal populations with unknown standard deviations; small samples are used, based on a variable t equal to the difference between the mean of the sample and the mean of the population divided by a result obtained by dividing the standard deviation of the sample by the square root of the number of individuals in the sample. ( http://www.answers.com/topic/t-test)
- DNA microarray
- A multiple assay procedure used in molecular biology. Thousands of short pieces of DNA call oglioneucleotides are placed on microscopic spots where they can then be exposed to probes that may or may not bind to them depending on sequence. They can be used to measure changes in DNA expression. (http://en.wikipedia.org/wiki/DNA_microarray)
- Protein expression
- A measurement of which proteins in a biological system have been translated and are thus present in a cell. This includes post-translationally modified proteins. ( http://www1.qiagen.com/about/Press/Glossary.aspx)
- Biomarkers
- Proteins found to have a biological significance for relevant biological conditions such as diseases. Biomarkers are highly important in drug discovery. (http://www1.qiagen.com/about/Press/Glossary.aspx)
Course Relevance
- The Utilization of the Significance Analysis of Microarray (SAM) analysis method in proteomic interpretation of data is a novel approach to solving the accuracy problems of LC data analysis. If false positive results can be identified accurately, then researchers could focus on the proteins that have been correctly characterized as changing in expression between two sets of conditions. This would allow for biomarkers to be found more readily.
Significance Analysis of Microarrays Applied to the Ionizing Radiation Response
[edit | edit source]Main Focus
The SAM analytical method is discussed as well as it validity for a given data set. The superiority of the analysis method over other methods is also discussed.
Summary
DNA Microarrays are capable of measuring the expression of thousands of genes within a single experiment. They are often used to identify any changes in expression of genes under a number of different conditions. Given that these experiments generate massive amounts of data, systems must be developed to analyze the data that is generated for any experimental significance. The authors of "Significance Analysis of Microarrays applied to the Ionizing Radiation Response" described a method for doing this known as SAM. This method provides a score for each gene based on its change in gene expression as compared to the standard deviation of repeat measurements. The system also provides an estimate for the false discovery rate. In the paper, the authors explain the algorithm behind how the SAM method arrives at these scores as well as the math behind determining the false discovery rate. SAM was tested on a data set that was collected by the authors and its validity was checked by Northern blots. The authors discuss SAM and its superiority over other methods of identifying experimental significance. These methods include identifying genes as having been significantly changed, if an R-fold change is observed, and declaring a significant change in expression based on whether or not an R-fold change was seen consistently between paired samples. SAM was shown to be superior to the other methods discussed3.
The algorithm behind SAM calculates the relative difference in gene expression based on permutation analysis of expression data and provides a score for these changes. It also calculates a false discovery rate. The algorithm used to calculate the score for each gene based on its change (if any) in gene expression is shown below:
New Terms
- SAM
- Significance Analysis of Microarray; A statistical analysis for determining whether or not changes in gene expression are experimentally significant. ( http://en.wikipedia.org/wiki/Significance_analysis_of_microarrays )
- False Discovery Rate (FDR)
- The percentage of genes incorrectly identified by chance. ( http://www-stat.stanford.edu/~tibs/SAM/pnassam.pdf )
- DNA Microarray
- DNA Microarrays are used in both molecular biology and medicine. They are made of a series of thousands of spots of DNA oligonucleotides arranged in a highly ordered fashion and can be used to measure changes in expression levels, to detect single nucleotide polymorphisms, and/or to resequence mutant genes. ( http://en.wikipedia.org/wiki/DNA_microarray )
- Northern Blot
- The northern blot is a technique typically used in microbiology that allows for the studying of gene expression by looking at RNA or isolated mRNA in the sample of interest. The technique allows for the observation of cellular control over structure and function by determining the particular gene expression levels during differentiation, morphogenesis, as well as abnormal or diseased conditions. ( http://en.wikipedia.org/wiki/Northern_blot )
- Ionizing
- The process of converting an atom or molecule into an ion by adding or removing charged particles. ( http://en.wikipedia.org/wiki/Ionization )
- Ion
- An atom or molecule that has either a positive or negative charge as a result of the addition or removal of an electron. ( http://en.wikipedia.org/wiki/Ion )
- R-fold Change
- This is a numerical values that evaluates the difference of group means. ( http://strimmerlab.org/software/st/html/diffmean.stat.html )
Course Relevance
- This method, which has previously been used to analyze DNA microarrays, is now being applied to proteomics data.
Websites Summarized
[edit | edit source]Mass Spectrometry Data Mining for Early Diagnosis and Prognosis of Stroke
[edit | edit source]Kalousis A. http://cui.unige.ch/AI-group/research/massspectrometry/massspectrometry.htm (28 March 2009)
Main Focus
The author describes the necessary signal conditioning steps in mass spectrometry spectral analysis before proteomics data are used in the classification of biomarkers.
Summary

From this website, the author describes the signal conditioning steps used to improve the integrity of mass spectrometry data and to reduce their high dimensionality. These steps include the uses of statistical techniques to exclude the influence of processing matrix, remove noises, minimize a data variation due to experiment conditions, and reduce the redundancy in the dataset.
Baseline removal was applied first to remove the data offset caused by the processing matrix usually used in the crystallization of protein sample for mass spectrometry. According to the website, the weighted quadratic fitting was used in this step. However, not all the effects of the matrix could be removed. Some remained and contributed in a form of noise in addition to the electrical noise from the machine itself. To alleviate the influence of noise, the denoising and smoothing techniques were used. The author reported that the wavelet decomposition together with a median filter was applied for this purpose. The spectral data was then normalized to dissociate the data from the experiment conditions. This process helped reduce a variation among data.
In the next step, a peak detection technique was used to remove the spatial redundancy of the data. Conceptually, it selected a mass-to-charge ratio value at the peak intensity in a specified range between two adjacent minima to represent the data in that range. Then, the selected spectral data were calibrated such that the data possessed the same spectral characteristic were clustered together. These clusters were referred to as the selected features, which were used by a machine learning algorithm in the classification of biomarkers.
New Terms
- Baseline removal (or baseline fitting)
- A method of removing an offset segment of spectral data so that the data can be further analyzed on the same basis. In general, the technique is to fit a curve to that offset segment and then subtract the values along the curve from the original data. As a result, the new spectral data will have a flat base. ( http://www.gb.nrao.edu/~rmaddale/140ft/unipops/unipops_7.html )
- Weighted quadratic fitting
- A curve fitting technique that uses a quadratic function to fit the weighted version of the original data. Usually, the weighting factor is derived from the error incurred from the regular quadratic fit. By accounting for this error, a better fit can be obtained. ( http://class.phys.psu.edu/p559/experiments/html/error.html )
- (Orthogonal) wavelet decomposition
- A signal analysis technique that uses a particular basis function, which is finite, to fit the signal and generates a series of time-varying coefficients. The decomposition yields a set of orthogonal signals. Each reflects a local change in the original signal at a given time. ( http://www.tideman.co.nz/Salalah/OrthWaveDecomp.html )
- Median filter
- A method of removing noise or data that has a distinctively large standard deviation for a given set of data. Consider a window of 2n + 1 data points. The median filter replaces the (n + 1)st data point with a median from this window. If the (n + 1)st data appears to be an out-of-range noise, it will be removed by this process. ( http://fourier.eng.hmc.edu/e161/lectures/smooth_sharpen/node3.html )
- Machine learning algorithm
- A sequence of instructions used by a computer such that the computer can adaptively improve its computational performance and efficiency in predicting the outcome based on the database it is collecting. ( http://en.wikipedia.org/wiki/Machine_learning )
Course Relevance
- This website addresses the problems associated with the data obtained from mass spectrometry and how to overcome those problems. It points out why the mass spectral data needs to be processed and how to do it before the data can be used in the analysis. This introduces another important aspect to the study of mass spectrometry in proteomics class.
Stanford School of Medicine Proteomics Center: Chu: Tibshirani Groups
[edit | edit source]Property of: Stanford University (3/29/09)
Main Focus
Gilbert Chu, M.D., Ph.D. and Robert Tibshirani, Ph.D. are attempting to create protein analysis tools/programs that will incorporated variable such as protein levels and post-translational modifications, and protein interactions in order to obtain more usable information from proteomic data.
Summary
This site offers information about the proteomic analysis research conducted by Gilbert Chu, M.D., Ph.D. and Robert Tibshirani, Ph.D. The site includes publications by Chu and Tibshirani (books and journal articles), as well as laboratory profiles for both professors. The overview provides concise information about the challenges and benefits of separating and analyzing proteomic data, and then merging it with gene expression data in order to obtain new information about biological systems. The software section on the website provides access to four different types of proteomic analysis software including Significance Analysis of Microarray(SAM), Prediction Analysis for Microarrays (PAM), laboratory statistical analysis for microarrays, and Peak Probability Contrast (PPC). The site also provides biographical background on the two professors as well as current research interests they are pursuing. These interest include elucidating a way to diagnose autoimmune diseases (such as systemic lupus erythematosus)from the analysis of proteins expressed in serum blood samples.

New Terms
- PPC
- Peak Probability Contrast - This is a class prediction software for protein MS data. It does this by obtaining a list of significant peaks where each class has an intensity level assigned to it. It can compare and contrast the intensity levels of the data to predict class and generate a false positive rate. Either raw spectra or extracted peaks can be input into this program.( http://proteomics.stanford.edu/chu/software.html )
- PAM
- Prediction Analysis for Microarrays - This is a program that performs sample classification from antibody reactivity data to provide a list of significant genes whose expression characterizes each sample group. The software can work with either cDNA or oligo microarrays. ( http://proteomics.stanford.edu/chu/software.html )
- cDNA
- complementary DNA - This is a single-stranded DNA that is reverse transcribed via the enzyme reverse transcriptase using a messenger RNA template. It is complimentary to the mRNA. (http://en.wikipedia.org/wiki/Complementary_DNA)
- Autoimmune disease
- A medical condition that occurs when the body's immune system mistakes its own normal tissue for foreign tissue and therefore launches an immune response attacking the tissue. There are over 80 known autoimmune disorders. (http://www.medterms.com/script/main/art.asp?articlekey=2402)
- SLE
- Systemic Lupus Erythematosus - This is an autoimmune disease more generally known as Lupus. The autoimmune response creates inflammation (both acute and chronic) in various tissues including heart, joints, skin, lungs, blood vessels, liver, kidneys, and nervous system. Most fatalities are due to renal failure and the 10yr survival rate is 80%. (http://www.medicinenet.com/systemic_lupus/article.htm)
Course Relevance
- The site provides information and access for multiple analysis software programs that may be used to analyze proteomic data
Introduction to FTICR/MS Technology
[edit | edit source]Wang B INTRODUCTION TO FTICR/MS TECHNOLOGY 2001
Main Focus
A brief introduction to the parts of a FTICR/MS machine and how it differs from other forms of MS.
Summary

Fourier transform mass spectrometry is a technique that has gained attention due to its ability to provide mass and resolution information from bio-molecules at levels that are higher than many forms of mass spectrometry. As an ion trapping form of mass spectroscapy it is unique in its technique on collecting these ions, it is for this reason that it is able to measure with far greater accuracy than other forms of mass spec. While the primary ideas for FTICR were first developed in the 1930s it was not until the late 1970s when Fourier transform techniques were adapted for ICR techniques. Since that time this technique has quickly increased in popularity and availability. While there are many types of FTMS machines they all contain four basic parts, which are necessary for their function. Primarily there is a magnet, which has a profound effect on the performance of the machine with stronger magnets yielding better results. It is for this reason that there is a constant movement toward stronger and stronger magnets. Secondly there is a cell that is used to store ions and is where they are detected and analyzed. Two types of cells can be utilized, cubic cells, composed of six magnetic plates arranged in a cube shape, hence its name, and open-ended cylindrical cells that perform in a manner similar to the cubic cells but utilizes six electrodes instead. The third feature is a vacuum system, a requirement to all mass spectrometers, but which is particularly important to the proper and accurate function to FTMS instruments, however this vacuum is only necessary when ions are detected in the trap. The final part of the system is the data system. The several components of this system, including a computer to monitor and analyze all of the other components and data have all improved in quality over the past ten years while their computer performance has also increased.
New Terms
- ion trap
- The ability to trap single to millions of atomic ions in good isolation from the outside world for a long period of time (http://jilawww.colorado.edu/pubs/thesis/king/ch2.pdf)
Course Relevance
- FTICR/MS is a popular tool in the feild of proteomics due to its ability to accurately identify peptide and protein products.
SAM: Significance Analysis of Microarrays
[edit | edit source]Property of: Stanford University (3/29/09)
Main Focus
This website provides information specifically about the SAM program developed at Standford University. It includes links to downloading and using the program as well as information regarding its background and algorithm.
Summary

This website is specifically dedicated to the SAM program. It includes information about how the program works (the algorithms behind it) as well as how it differs from other analysis programs. It provides a link to the instruction manual. They give a list of features for the program as well as links to other extras that may be used alongside it (such as an Excel add-in). A FAQ section is also available to answer the most popular questions. There are links to download the SAM program in addition to information about how to get licensing (if it is necessary). Most users can download the program simply by registering at the appropriate site (http://www-stat-class.stanford.edu/~tibs/clickwrap/sam.html), however, if SAM is going to be for commercial use, more formal licensing is required. Additionally, the site keeps everyone up to date about any changes they make to SAM as well as when new versions come out.
New Terms
- PAM
- Prediction Analysis of Microarrays; Another system available through the SAM website that is used for class prediction and survival analysis for gene expression as well as data mining. The system performs sample classification from gene expression data. ( http://www-stat.stanford.edu/~tibs/PAM/index.html )
- Algorithm
- A finite sequence of instructions usually in the form of an explicit, step-by-step procedure for solving a problem. It is often used for calculation and data processing. ( http://en.wikipedia.org/wiki/Algorithm )
- Two Class (Unpaired) Groups
- Two sets of measurements, in which the experiment units are all different in the two groups. ( http://www-stat.stanford.edu/~tibs/SAM/sam.pdf )
- Multiclass
- There are more than two groups, each containing different experimental units. ( http://www-stat.stanford.edu/~tibs/SAM/sam.pdf )
- Normalization
- This is a way of systematically making sure that a database is able to carry out general-purpose queries and that these queries don't result in undesirable results by either insertion, updating, or deletion of any anomalies that can results in loss of the quality of the data. ( http://en.wikipedia.org/wiki/Database_normalization )
Course Relevance
The site offers further information about the SAM program, which is now being utilized in proteomics studies. It also provides access to downloading SAM.
An (Opinionated) Guide to Microarray Data Analysis
[edit | edit source]http://www.bea.ki.se/staff/reimers/Web.Pages/Microarray.Home.htm (3/29/09)
Main Focus
Overall, this website provides information about common problems in the analysis of microarray data and presents suggestions on how to resolve these problems. Some of these solutions take the form of normalization techniques.

Summary
The website is organized into eleven total sections—Experimental design, Distributions and transforms, Approaches to Normalization, Quality Control of spotted arrays, Normalization of spotted arrays, Quality control, Normalization, Estimates of Abundance (methods for combining data from multiple probes to get single estimates), Graphics, Clustering, and Statistical Significance—that explain some of the different techniques people use to solve some of the problems that arise when analyzing microarray data. The first three sections talk about problems that arise in all types of microarray analysis. The website gives a brief explanation of the concepts being covered and then proceeds to talk about their strong and weak points. The next five sections then talk about these concepts specifically for two-color cDNA spotted microarrays and for the Affymetrix arrays respectively. In the Quality Control sections the website goes over some basic controls that should be kept in mind before running the experiment just to cut down on systematic bias. While normalization of the data will account for some of the bias, it won't necessarily catch everything, so it is best to avoid introducing bias if possible. The last section discusses different statistical methods that are appropriate for microarray data and the other two sections discuss different ways that the data is then visualized for human analysis.
New Terms
- p-value
- probability that given the null hypothesis is true, the results obtained from the experiment make sense (http://www.childrensmercy.org/stats/definitions/pvalue.htm)
- Bonferroni correction
- a method that is used to keep results arising from multiple comparison tests from being incorrectly deemed statistically significant (http://www.utdallas.edu/~herve/Abdi-Bonferroni2007-pretty.pdf)
- Sidak correction
- another correction method that is less stringent than the Bonferroni correction allowing a greater chance for something to be considered statistically significant (http://www.utdallas.edu/~herve/Abdi-Bonferroni2007-pretty.pdf)
- Lowess curve (locally weighted linear regression curve)
- a smooth curve that's plotted through the points and is calculated by using locally weighted linear regression over the values (http://www.itl.nist.gov/div898/software/dataplot/refman1/ch3/lowess_s.pdf)
- t-test
- a statistical hypothesis test that follows the Student's t distribution if the null hypothesis is true (http://www.socialresearchmethods.net/kb/stat_t.php)
Course Relevance
Most of the normalization techniques used for Proteomics were first found useful for microarray data, so learning about problems that arise in microarray data analysis can be useful when doing experimental design.
Notes
[edit | edit source]- ↑ Ge G, Wong GW. "Classification of premalignant pancreatic cancer mass-spectrometry data using decision tree ensembles" BMC Bioinformatics 9:275 (2008).
- ↑ Stephen J. Callister; et al. (2006). "Normalization Approaches for Removing Systematic Biases Associated with Mass Spectrometry and Label-Free Proteomics". J Proteome Res. 5 (2): 227–286. doi:10.1021/pr050300l. PMID 16457593.
{{cite journal}}:|access-date=requires|url=(help); Explicit use of et al. in:|author=(help)
References
[edit | edit source]1 - Ge G, Wong GW. "Classification of premalignant pancreatic cancer mass-spectrometry data using decision tree ensembles" BMC Bioinformatics 9:275 (2008).
2 - Stephen J. Callister et al. (2006). "Normalization Approaches for Removing Systematic Biases Associated with Mass Spectrometry and Label-Free Proteomics" J Proteome Res. 5(2): 227–286. (http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1992440)
3 - Tusher, V. G., R. Tibshirani, et al. "Significance Analysis of Microarrays Applied to the Ionizing Radiation Response." PNAS 98:5116–5121 (2001).
4 - Kalousis A. "Mass spectrometry data mining for early diagnosis and prognosis of stroke" http://cui.unige.ch/AI-group/research/massspectrometry/massspectrometry.htm (28 March 2009)
5 - Zimmer JSD, Monroe ME, Qian WJ, Smith RD Mass Spectrom Rev. 25(3):450-482 (2006) Advances in Proteomics Data Analysis and Display Using an Accurate Mass Time Tag Approach (http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1829209)
6 - Li Q, Roxas BAP. "Significance Analysis of Microarray for Relative Quantitation of LC/MS Data in Proteomics" BMC Bioinformatics 9:187 (2008)