Next Generation Sequencing (NGS)/Print version
→
Introduction
ABOUT THIS BOOK
- The first four chapters are general introductions to broad concepts of bioinformatics and NGS in particular. They are 'required pre-requisites', and will be referred to in the rest of the book:
- In the Introduction, we give a nearly complete overview of the field, starting with sequencing technologies, their properties, strengths and weaknesses, covering the various biological processes they can assay, and finishing with a section on common sequencing terminology. Finally we finish with an overview of a typical sequencing workflow.
- In Big Data we deal with some of the (perhaps unexpected) difficulties that arise when dealing with typical volumes of NGS data. From shipping hard drives around the world, to the amount of memory you'll need in your computer to assemble the data when they arrive, these issues often take novices by surprise. We'll get into the file formats, archives, and algorithms that have been developed to deal with these problems.
- In Bioinformatics from the outside we will discuss the interfaces used by bioinformaticians. We will present the command line with its text interface and blinking cursor, but also more user friendly graphical user interfaces (GUIs) which were developed specially for bioinformatics pipelines.
- In Pre-processing we will discuss the best practices of controlling the quality of a NGS dataset, and cleaning out low quality data.
- The next five chapters describe the analyses which can be done using a reference genome sequence, assuming one is available:
- In Alignment we will discuss how to map a set of reads to a reference dataset.
- In DNA Variation we will describe how to call variants (either SNVs, CNVs or breakends) using mapped reads.
- In RNA we will explain how to determine exons, isoforms and gene expression levels from mapped RNA-seq reads.
- In Epigenetics we will describe pull down assays which are used to determine epigenetic traits such as histone or CpG methylation.
- In Chromatin structure we will discuss technologies used to determine the structure of the chromatin, e.g. the placement of the histones or the physical proximity of different chromosomal regions when the DNA lies in the nucleus.
- Finally the last two chapters will describe analyses in the absence of a reference genome:
- De novo assembly will describe how to assemble a genome from NGS reads.
- De novo RNA assembly will explain how to assemble a transcriptome from NGS reads only.
Introduction
Platforms and Technologies
NGS platforms employ different technologies to decode the identity of nucleotides in DNA, or detect covalent modifications such as methylation on the nucleotides.
NGS platforms evolve quickly. Usually, new technologies & platforms are announced at the Advances in Genome Biology & Technology (AGBT) conference [1]
For educational purposes, some reviews of NGS platforms published in 2011 [2]. Read more about the sequencing technologies here
File format and terminology
FASTA
The FASTA format, generally indicated with the suffix .fa or .fasta, is a straightforward, human readable format. Normally, each file consists of a set of sequences, where each sequence is represented by a one line header, starting with the '>' character, followed by the corresponding nucleotide sequence, in multiple lines of regular width (generally 60 or 80 characters wide). In practice, some tools may produce a sequence with a header and a single long line of sequence. For more detailed information see the FASTA Wikipedia page.
FASTQ
FASTQ is a text file format (human readable) that provides 4 lines of data per sequence.
- Sequence identifier
- The sequence
- Comments
- Quality scores
FASTQ format is commonly used to store sequencing reads, in particular from Illumina and Ion Torrent platforms.
Paired-end reads may be stored either in one FASTQ file (alternating) or in two different FASTQ files. Paired-end reads may have sequence identifiers ended by "/1" and "/2" respectively.
Example FASTQ entry for one Illumina read:
@EAS20_8_6_1_3_1914/1 CGCGTAACAAAAGTGTCTATAATCACGGCAGAAAAGTCCACATTGATTATTTGCACGGCGTCACACTTTGCTATGCCATAGCATTTTTATCCATAAGATT + HHHHHHHHHFHGGHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHHHHGHHHGHHHGHIHHHHHHHHHHHHHHHGCHHHHFHHHHHHHGGGCFHBFBCCF
Generally a FASTQ file is stored in files with the suffix .fq or .fastq using Gzip file compression indicated by the suffix .gz or .gzip.
For more detailed information see the FASTQ Wikipedia page.
SFF
SFF is a binary file format used to encode sequencing reads from the 454 platform.
http://en.wikipedia.org/wiki/Standard_Flowgram_Format
SAM/BAM
File formats used to encode short reads alignment. See Next_Generation_Sequencing_(NGS)/Alignment for more information.
FASTG
FASTG is an emerging file format for genome assemblies that take ambiguities into account. FASTG is like FASTA, but the G stands for ‘graph’.
VCF
The Variant Call Format (VCF) is a specification used in bioinformatics for storing gene sequence variations. See [1] for more information.
Read lengths
As of Feb 2013, the read-length of second generation sequencing platforms are shorter than conventional Sanger sequencing, creating challenges in reads mapping and assembly.
- The most well used Illumina platforms can produce read-length up to 250bp. In practice, ~100bp is mostly accessible to researchers worldwide.
- Ion Torrent: Varies, typically peak at 400bp
- SOLiD: 50-75bp
Paired-/Single-ends
- Single-end reads means the sequence fragment are sequenced from 1 direction only.
- In paired-end sequencing, a single fragment are sequenced from both 5' and 3' end, giving rise to forward and reverse read. The sequenced fragments could be separated by a certain bases (inner insert size) or can be overlapping, giving rise to a contiguous longer single-end fragment after merging. The uses of paired-end reads can improve the accuracy of reads mapping onto a reference genome. The typical fragment size (external inserts size) is 200bp to 500bp
Mate-pairs
Mate-pair is different from paired-end in the sense of how the sequence library is made. In mate-pair sequencing, 2-5kb fragments are selected and sequenced from both ends, thus giving information how nucleotides far apart are linked together. Mate-pairs are more ideal for studying genomic structural rearrangement and help de novo genome assembly. They also facilitate sensitive structural variant (SV) detection across a widened SV size-spectrum and in repetitive areas of the genome.
Colorspace
Colorspace is a 2-base encoding system commercialized by Life Tech and used in SOLiD platforms. Technology overview is described here.
Quality scores
Quality score is an indication of probability of the base call being incorrect. Quality score is used in the FASTQ format.
Various encoding schemes are available, including, most commonly, [Phred quality scores].
Error profiles & Sequencing biases
Uses of NGS
DNA
To find mutations from tumor cells .
RNA
To reconstruct transcriptome (genome-based or de novo) using reverse transcription so that researchers can count how many reads align onto annotated parts of the transcriptome. This is used to compare gene expression in samples that are dramatically different from each other and to build biochemical pathways of an organism.
ChIP
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations.
Chromatin structure
General NGS Workflow Overview
References
Big Data
Big Data
Data Deluge
The first problem you face is probably the large size of the NGS FASTQ files - the "data deluge" problem. You no longer only have to deal with microplate readings, or digitalized gel photos; the size of NGS data can be huge. For example, compressed FASTQ files from a 60x human whole genome sequencing can still require 200Gb. A small project with 10–20 whole genome sequencing (WGS) samples can generate ~4TB of raw data. Even these estimates do not include the disk space required for downstream analysis.
Storing data
Referenced from a post from BioStars[1]:
- Very high end: enterprise cluster and SAN.
- High end: Two mirrored servers in separate buildings or Cloud.
- Typical: External hard drives and/or NAS with raid-5/6
Moving data
Moving data between collaborators is also non-trivial. For RNA-Seq samples, FTP may suffice, but for WGS data, shipping hard drives may be the only solution.
Externalizing compute requirements from the research group
It is difficult for a single lab to maintain sufficient computing facilities. A single lab will probably own some basic computing hardware; however, many tasks will have huge computational demands (e.g. memory for de novo genome assembly) that require them to be performed elsewhere. An institution / core facility may host a centralized cluster. Alternatively, one might consider doing the task on the cloud.
- NIH maintains a centralized computing cluster called Biowulf.
- Bioinformatics cloud computing is suggested.[2][3] EBI has adopted a cloud-based platform called Helix Nebula.[4]
References
- ↑ Wo, H. (24 March 2011). "Question: Huge Ngs Data Storage And Transferring". Biostars. Biostar Genomics, LLC. Retrieved 28 April 2016.
- ↑ Akhlaghpour, H. (3 July 2012). "Genomic Analysis in the Cloud". YouTube. Google. Retrieved 28 April 2016.
- ↑ Schadt, E.E.; Linderman, M.D.; Sorenson, J.; Lee, L.; Nolan, G.P. (2010). "Computational solutions to large-scale data management and analysis". Nature Reviews Genetics. 11 (9): 647–57. doi:10.1038/nrg2857. PMC 3124937. PMID 20717155.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Lueck, R. (16 January 2013). "Big data and HPC on-demand: Large-scale genome analysis on Helix Nebula – the Science Cloud" (PDF). Trust-IT Services. Retrieved 28 April 2016.
Bioinformatics from the outside
Bioinformatics from the outside
- For an in-depth introduction to UNIX, see the Guide to Unix or A Quick Introduction to Unix.
Unix command line: History
The first version of Unix was developed by Bell Labs (part of AT&T) in 1969, making it more than forty years old. Its roots go back to when computers were large and rare, time on them very expensive and shared between many users. Unix was developed so as to allow multiple users to work simultaneously. Unix actually grew out of a desire to play a game called Space Travel and the features that made it an operating system were incidental. Initially it only supported one user and the name Unix, originally UNICS, is a pun on MULTICS, a multi-user system available at the time.
While this might seem strange and unnecessary in a world where everyone has their own laptop, computing is again moving back to remote central services with many users. The compute power required for mapping next-generation sequencing data or de novo assembly is beyond what is available or desirable to have sitting on your lap. In many ways, the “cloud” (or whatever has replaced it by the time you read this) requires ways of working that have more in common with traditional Unix machines than the personal computing emphasised by Windows and Apple Macintosh.
USA federal monopoly law prevented AT&T from commercialising Unix but interest in using it increased outside of Bell Labs and eventually they decided to give it away freely, including the source code, which allowed other institutions to modify it. Perhaps the most important of these institutions was the University of Berkeley. (A significant proportion of Mac OS X has its roots in the Berkeley Standard Distribution (BSD) that distributed a set of tools to make Unix more useful and made changes that significantly increased performance.) The involvement of several universities in its development meant Unix was ideally placed when the internet was created and many of the fundamental technologies were developed and tested using Unix machines. Again, these improvements were given away freely. Some of the code was repurposed to provide networking for early versions of Windows and even today several utilities in Windows Vista incorporate Berkeley code.
As well as being a key part in the development of the early internet, a Unix machine was also the first web server, a NeXT cube. NeXT was an early attempt to make a Unix machine for desktop use. Extremely advanced for its time but also very expensive, it never really caught on outside of the finance industry. Apple eventually bought NeXT, its operating system becoming OS X, and this heritage can still be seen in its programming interfaces. Apple is now the largest manufacturer of Unix machines; every Apple computer, the iPhone, and most recent iPods have a Unix base underneath their facade.
By the early 90s Unix became increasingly commercially important. This inevitably lead to legal trouble: with so many people giving away improvements freely and having them integrated into the system, who actually owned it? The legal trouble cast uncertainty over the freely available Unix versions, creating an opening for another free operating system.
The vacuum was filled by Linux, a freely available computer operating system similar to Unix started by Linus Torvalds in 1991 as a hobby. More correctly, Linux is just the kernel, the central program from which all others are run. Many more tools in addition to this are required to make an operating system. These tools are provided by the GNU project.[1]
Importantly, Linux was written from scratch and did not contain any of the original Unix code and so was free of legal doubt. Coinciding with the penetration of the internet onto university campuses, and the availability of cheap but sufficiently powerful personal computers, Linux rapidly matured with over one hundred developers collaborating over the internet within two years. The real advances driving Linux were social rather than technological, disparate volunteers donating time on the understanding that, in return for giving their work away freely, anything based on their work is also given away freely and so they in turn benefit from improvements.
The idea that underpins this sharing and ensures that nobody can profit from anyone else's work without sharing is “copyleft”, described in a simple legal document called the GNU General Public Licence,[2] which turns the notion of copyright on its head. (It should be noted that the GNU project, and the philosophy behind it, predate Linux by almost a decade.) Today, Linux has become the dominant free Unix-like operating system with millions of users and support from many large companies.
Getting and installing Ubuntu
Here we describe the Ubuntu distribution (packaging) of Linux, which is one of the most widely used, but all the examples are fairly generic and should work with most Linux, Unix, and Mac OS X computers. There are many different guides on the web about how to install Ubuntu but we recommend installing it as a virtual machine on your current computer.
The Ubuntu Linux distribution is generally easy to use and it is updated (for free) every six months. The examples and versions used here are for version of Ubuntu is 11.10, named after its release date in October 2011, and also known as “Oneiric Ocelot”; the next (most current) version, 12.04 or “Precise Pangolin” was released in April 2012 and is designated a Long Term Support (LTS) edition, meaning that it will be receive fixes and maintenance upgrades for five years before being retired, and is the best option if you don't want to be regularly upgrading your system.
Acclimatisation
A significant effort has been undertaken to make Ubuntu easy to use, so even novice computer users should have little trouble using it. There are quite a few tutorials available for users new to Ubuntu. The official material is available[3] but a quick search on the web will locate much more. In addition, there is a lot of documentation installed on the machine itself. You can access this by moving the mouse towards Ubuntu Desktop at the top left of the screen and clicking on the help menu that appears. In general, the name of the program you are currently using is displayed at the top-left of the screen and moving the mouse to top of the screen will reveal the programs menus in a similar fashion to how they are displayed on the Mac (although, confusingly, some programs display their menus within their own window rather like a Windows computer).
An alternative way to get help is to click on the circular symbol (a stylised picture of three people holding hands) at the top left of the screen and type help in the search box that appears. For want of a better name, we will refer to the people-holding-hands button as the Ubuntu button although the help text that appears describes it as “Dash home”.
Ubuntu comes free with many tools, including web browsers, file managers, word processors, etc. There are free equivalents available for most of the everyday software people use, and you can browse what is available by clicking on the Ubuntu Software Centre, whose icon at the left of the screen looks like a paper shopping bag full of goodies. The Ubuntu Software Centre is just a starting point and there are many other sources available, both of prepackaged software specifically for Ubuntu, and source code that will require compiling. Search the web for “Ubuntu software repositories” for more information on obtaining additional software.
While there are explicit key combinations for copy and pasting text, just like on Windows or Mac, control-c and control-v in Ubuntu, this convention is not respected by all programs. Unix has traditionally been more mouse centred with the left mouse button used to highlight text and the middle button used to copy it. You may find yourself accidentally doing this occasionally if you are not used to using the middle mouse button. Starting applications from icons, opening folders, etc... only requires a single click, rather than the double click required on Windows, making the action of pressing buttons and selecting things from menus more consistent with each other. Accidentally double clicking will generally result in an action being done twice, not normally a bad thing but it does mean that impatient users can quickly find their desktop covered in windows.
Perhaps the most important difference you are likely to encounter on a daily basis is that the names of files and directories are case sensitive: README.txt, readme.txt and readme.TXT all refer to different files. This is different from both Windows and Mac OS X, where upper and lower-case characters are preserved in the name but the file can be referred to using any case. (Despite the Unix heritage of OS X, Apple chose this behaviour to maintain compatibility with earlier versions of the Mac operating system)
Fetching the examples
There are many examples in this tutorial to be tried, enclosed in boxes like the one below, which explains the format of the examples. The example below shows how to automatically download and unpack the file ready for use.
Basics
The command line
While Ubuntu has all the graphical tools you might expect in a modern operating system, so new users rarely need to deal with its Unix foundations, we will be working with the command-line. An obvious question is, why is the command-line still the main way of interacting with Unix or, more relevantly, why we are making you use it? Part of the answer to the first question is that the origins of Unix predate the development of graphical interfaces and this is what all the tools and programs have evolved from. The reason the command-line remains popular is that it is an extremely efficient way to interact with the computer: once you want to do something complex enough that there isn't a handy button for it, graphical interfaces force you to go through many menus and manually perform a task that could have been automated. Alternatively, you must resort to some form of programming (Mac OS X Automator, Microsoft Office macros, etc) which is the functional equivalent of using the command line.
Unix is built around many little tools designed to work together. Each program does one task and returns its output in a form easily understood by other programs. These properties allow simple programs to be combined together to produce complex results, rather like building something out of Lego bricks. The forward to the 1978 report in the Bell System Technical Journal[4] describes the Unix philosophy as:
"(i) Make each program do one thing well. To do a new job, build afresh rather than complicate old programs by adding new features.
(ii) Expect the output of every program to become the input to another, as yet unknown, program. Don't clutter output with extraneous information. Avoid stringently columnar or binary input formats. Don't insist on interactive input.
(iii) Design and build software, even operating systems, to be tried early, ideally within weeks. Don't hesitate to throw away the clumsy parts and rebuild them.
(iv) Use tools in preference to unskilled help to lighten a programming task, even if you have to detour to build the tools and expect to throw some of them out after you've finished using them."
The rest of this tutorial will be based using the command-line through a “terminal”. This terminology dates back to the early days of Unix when there would be many “terminals”, basically a simple screen and keyboard, connected to a central computer. The terminal program can be found by clicking on the Ubuntu button and typing terminal in the search box, as shown in Illustration 1. You can also easily access the terminal using just the keyboard, by pressing control-alt-T. Once open, the text size can be changed using the View/Zoom menu options or the font changed entirely using the Edit/Profile Preferences menu option.
While we are using Linux during the workshop, you may not have access to a machine later or may not wish to use Linux exclusively on your computer. While you could install Linux as 'dual-boot' on your computer, or run it in a virtual machine (A Virtual Machine (VM) is a program on your computer that acts like another computer and can run other operating systems. Several VM's are available, VirtualBox http://www.virtualbox.org/ is free and regularly updated), the knowledge of the command-line is fairly transferable between platforms. Mac OS X also has a command-line hidden away (/Applications/Utilities/Terminal) and, with a small number of eccentricities, everything that works on the Linux command-line should work for OS X. Windows has its own incompatible version of a command-line but Cygwin http://www.cygwin.com/ can be installed and provides an entire Unix-like environment within Windows.

At the beginning of the command-line is the command prompt, showing that the computer is ready to accept commands. The prompt is text of the form user@computer:directory$. Figure 2 has a user called tim in the directory ~ on a computer called coffee-grinder. Having all this information is handy when you are working with multiple remote computers at the same time. The prompt is configurable and may vary between computers; you may notice later that other prompts are slightly different. Some basic commands are shown in Table 1; try typing them at the command-line and press return after the command to tell the computer to run the command.
Files and directories
All files in Unix are arranged in a tree-like structure: directories are represented as branches leading from a single trunk (the “root”) and may, in turn, have other branches leading from them (directories inside directories) and individual files are the leaves of the tree. The tree structure is similar to that of every other common operating system and most file browsers can display the filesystem in a tree-like fashion, for example: part of the filesystem for an Ubuntu Linux computer is displayed in Figure 4.

Where Unix differs from other operating systems is that the filesystem is used much more for organising different types of files. The essential system programs are all in /bin and their shared code (libraries) are in /lib; similarly user programs are in/usr/bin, with libraries in /usr/lib and manual pages in /usr/share/man.
There are two different ways of specifying the location of a file or directory in the tree: the absolute path and the relative path from where we currently are in the filesystem (the current working directory). An absolute path is one that starts at the root and does not depend on the location of the current working directory. Starting with a / to signify the root, the absolute path describes all the directories (branches) we must follow to get to the file in question. each directory name is separated by a /.
For example, home/user/Music/TheKinks/SunnyAfternoon.mp3 refers to the file SunnyAfternoon.mp3 inside the directory TheKinks, which is inside the directory Music, which is inside the user's directory, which is inside on the directory home, which is connected to the root. If you are familiar with Microsoft Windows, you might notice that the path separator is different. Unix-based systems use a forward-slash (/) rather than the backward-slash (\) used on Windows. You may have noticed that the paths of web pages are also separated by forward-slashes, revealing their Unix origins as a path to a file on a remote machine.
For convenience, a few directories have special symbols that are synonyms for them and the most common of these are listed in Figure 5. Most of these have a special meaning when at the beginning of a path otherwise they are just a symbol. For example dir/~/ is the directory ~ inside the directory dir in the current directory, whereas ~/dir/ is the directory dir inside the home directory (usually /home/user on Linux, /Users/user on Mac OS X). In both cases the '/' symbols are separators rather than the root directory.
The current location, the working directory, can be displayed at the command-line using the pwd command. Rather than referring to a file by its absolute path, we can refer it by using a path relative to where we are: a file in the current directory can be referred to by its name, a file in a directory inside our working directory can be referred to by directory/filename (and so on for files inside of directories inside of directories inside of our working directory, etc...). Note that these paths are very similar to how we describe absolute paths except that they do not start with /; absolute paths are relative paths relative to the root (alternatively we could read the initial / as “goto root” and consider them to be relative paths). As shown in Figure 5, the directory above the current directory can be referred to as .. so, if the working directory is /home/user, then the root directory can be referred to as ../.. (go up one directory, then go up another directory). The symbol .. can be freely mixed into paths: the directory examples below the current directory could have path examples/../examples/../examples (needless to say, simply using just examples is recommended).
Commands
Commands are just programs elsewhere on the computer and entering their name on the command-line runs them. Commands have a predicable format:
command -flags target
The command is the name of the program to run, the (optional) flags modify its behaviour and the target is what the command is to operate on, often the name of a file. Many commands require neither flags nor target but Unix tools are generally extremely configurable and even simple commands like date (some utilities also have parodies, see ddate or sl for example) have many optional flags to change the format of their output.
As mentioned in Files and directories, there are special directories to contain executable programs and programs within them can be run by typing their name at the command-line. The reason you can run the programs in these directories simply by typing their names is that the operating system knows to look in those directories for programs. In general you will not have permission to place files in these directories and experienced Unix users create their own, normally ~/bin/ ,to place programs they use frequently. Creating this directory does not make it special; you still have to tell the operating system to go look for programs there as well. The operating system has a variable, $PATH, which is a list of directories in which the computer looks for programs. To add a directory to that list, use the command "export PATH=~/bin:$PATH" where "~/bin" is the directory you want to add. This command is often added to the file ~/.bashrc, which is a list of commands to be run automatically every time a new terminal is opened. If a program is not in a special directory, you cannot run it just by typing its name because the computer doesn't know where to find it. This is true even if the program is in the current directory. Programs which are not in special directories can still be run, but you have to include the path to where it can be found. If the program is in your current working directory, this can be as simple as typing ./program (program is in current directory). If the program is elsewhere just type the absolute or relative path to were it is. You can always use the command-line's autocompletion features (see “tab-completion” below) to reduce the amount of typing needed. In order to alleviate the need to type paths to commonly-used programs, it is a good idea to add their paths to the PATH variable in ~/.bashrc.
One thing you'll quickly discover is that the mouse does not move the cursor in the terminal. The terminal interface predates the popularity of mice by decades and alternative methods of efficiently moving around and editing have been developed. There are keyboard short-cuts defined for most common operations, and a few of these are listed in Figure 6. Probably the most useful shortcut is the tab key. It can be used to complete command names and paths in the filesystem (called 'tab-completion'). Pressing tab once will complete a path up to the first ambiguity encountered and pressing again gives a list of possible completions (you can type the next letter or so of the one you want and press tab again to attempt further auto-completion).

A record is kept of the commands you have entered, and the history command can be used to list them so you can refer back to what you did earlier. The history can also be searched: Control-r starts a search and the computer will match against your history as you type; typing enter accepts the current line, typing Control-r again goes to the next match and Control-g cancels the search. History can also be referred to by entry number, listed using the history command: entering !n on the command-line will repeat history entry n, entering !! will repeat the last command.
There are many commands, often quite terse, for manipulating files and a few of the more useful of these are shown in Table 4. Many of the commands for Unix have short names, often only two or three letters, so errors typing can easily have unintended and severe consequences! Be careful what you enter, because Unix rarely gives you a second chance to correct mistakes. Some Unix machines have the sl command to encourage accurate typing.
Figure 7 shows few commands for manipulating files and brief explanations.
On the Unix command line, some symbols can have special meanings. A slash, '/', indicates the end of a directory name, an asterisk, '*' is a wildcard, etc. However, there are many circumstances when it is preferable for symbols not to have a special meaning, the most common example being when the file name contains a space (a space is a special character in the sense that it is interpreted as a break between command-line options). The character in question can be “escaped” by prefixing it with a '\' to remove its special meaning so, for example: / is the root directory but \/ is a file called '/'.
Files beginning with a . character are hidden by default and will not appear in the output of ls or equivalent (or in the file browser when you're using a graphical user interface). Generally, hidden files are those used directly by the computer or programs, containing configuration information not intended for the average user to understand or use.
Reading and writing permission
All files and directories have a set of permissions associated with them, describing who is allowed to read or write that file. There are three basic permissions: read r, write w, and execute x. The meanings of read and write are fairly obvious, but execute has two meanings depending on context. For normal files, execute permission is used on files with executable code (i.e. programs) to give users permission to run that program. For a directory, x permission allows a user to open that directory and see the files it contains. There are three categories of user: owner u (generally the user who created the file), group g (the group of users that the owner belongs to), and other o (everyone else). The permissions for each file are described as a string of nine characters, three for each user category. The three positions assigned to each user category correspond to the three types of permissions ('r,w, and x, in that order). If that user category has a given permission, the appropriate letter will appear. If not, the letter will be replaced with a dash '-'. For example, if a user category has permission to read and execute a file, but not write it, their triplet will look like r-x. The permission string rwxr-x--- means that the owner has permission to read, write or execute, users in the same group have read and execute permission and other users have no permissions.
The owner of a file can change its permissions. Some programs will do this automatically if they are being run by the file's owner, giving the impression that the permissions have been ignored. Running rm -f is the most common time a user will run into this behaviour: by default rm will prompt to remove write-protected files (i.e. files you don't have permission to write) but the -f (force) flag turns tells it not to bother asking and just remove the file.
Dealing with multiple files
Often, especially when running scripts or organising files, you will need to be dealing with multiple files at once. Rather than typing each file name out explicitly, we can give the computer a pattern instead of a filename. All filenames are checked against the pattern and the computer automatically generates a list of all the matching files to use when running the command. Patterns are created using symbols that have a special meaning. For example: * means match anything (or nothing), so a*b is a pattern that matches any filename beginning with a and ending with b including the file ab. Figure 8 contains a list of special symbols useful for constructing patterns.
As mentioned above, pattern matching occurs before a command is run and the pattern is replaced by the list of matches. The command never sees the pattern, just the results of the match.
Running multiple programs
From early on in its development, Unix was designed to run multiple programs simultaneously on remote machines and support for this is integrated into the command-line. Jobs (scripts, programs, or other fairly self-contained things running from the command line) can be divided into two types, foreground jobs and background jobs, based on how they affect the terminal. A foreground job temporarily replaces the command-line and you cannot enter new commands until it has finished, whereas a background job runs independently and allows you to continue with other tasks. Only foreground jobs receive input from the keyboard, so interactive programs like PAUP* should be run as foreground (although you could set up a compute intensive analysis, background it and continue with other tasks while it is running. Later, when the calculations have finished, the program can be made foreground again so interaction can continue). Although background jobs leave your command-line mostly free to do other things, they do send their output to the terminal you launched them from, so you might see it popping up in the middle of another task, which can be confusing. If you are running multiple background jobs, their output will be interleaved based on when it was produced, with no indication of which program produced the output.
As hinted in Figure 9, there is a difference between a job and a process. A process is a single program running on the machine, and each process is uniquely numbered with a pid (process ID). You can list all the processes you are running, including the command-line itself (generally called bash, but in some unix distributions it may be zsh or tcsh) using ps (or ps -a if you want to see what all the other users of the machine are doing). The command-line itself is just a process (program) running on the computer, albeit one specially designed for starting, stopping and manipulating other processes. Processes are the fundamental method of keeping track of what is running on the computer. Jobs, on the other hand, are things entered on the command-line and many include several programs logically connected together by pipes (see In, out and pipes
for details) to achieve a task.

The command-line
splits the jobs into several processes and runs them, possibly simultaneously. See illustrative example in Figure 10.
In, out and pipes
Where possible, Unix commands behave sort of like filters, or the mathematical concept of a function: they read from input, manipulate that input, and write the output. This might sound trivial, tautologous even, but it enables simple commands to be combined to produce complex results. Every command reads from stdin (short for standard in) and writes to stdout (short for standard out). By default stdin is whatever gets typed in the terminal from and keyboard, and stdout is connected to the current command-line, so results are displayed on the screen. stdout can easily be redirected to a file instead by using the greater-than operator, >. > filename redirects stdout to the file specified for later perusal. By chaining many simple commands together, complex transformations of the input can be achieved. The following is an advanced example, showing how a complex output can be achieved using a series of smaller steps. You may not yet have sufficient understanding of the shell to follow everything in this example but try to work through it and see what each step is doing. The main pages for each command (see Getting help) might be useful.
Compression
The aim of compression is to make files smaller, which is useful for both saving disk space and making it quicker to send files over the internet. Some types of programs that send data over the internet have the ability to transparently compress files before sending and uncompress at the other end. Some web servers implement this but the most important example for us are scp and sftp (two command-line programs used to transfer files over networks) which can each be given the -C option to request compression.
Simply put, compression programs look for frequently repeated patterns in the file and remove this redundancy in a manner that can be undone later. Text files tend to compress very well, with 100MB worth of Wikipedia being compressable into less than 16MB (See The Hutter prize http://prize.hutter1.net/), and, in particular, biological sequences tend to be very compressible since the size of the alphabet of nucleotides or amino acids is small.
The two most common tools for compressing files are gzip and bzip2, with their respective tools for uncompressing being gunzip and bunzip2. gzip is the de-facto standard; bzip2 tends to produce smaller files but takes longer to compress them. On the Windows platform, the Zip (often known as WinZip (http://www.winzip.com/)) compression method is favoured and many Unix platforms provide zip and unzip tools to deal with these files. Non-Linux Unix platforms, like Mac OS X for example, have older tools called compress and uncompress that are rarely used any more. Support for compress 'd files on Linux can be patchy and unreliable. For example, a machine one author has access to has a compress manual page but no actual tool installed.
A final method to be aware of, that is becoming more popular, is 7-zip (7za). 7-zip can produce smaller files than all the above methods, again at the expense of taking longer to compress. A list of file suffixes that can be used to identify what files are compressed using what method is provided in Figure 11.
Compression works better if files are combined and then compressed together, rather than compressing them individually, since this allows the compression program to spot repeated patterns between the files. On Unix, the process of packing/unpacking several files into / from a single file has been historically separate from the process of the compression, in keeping with the philosophy of having little tools that do one thing well. The Unix tool for packing and unpacking files is tar “Tape Archiver”, the odd name because its heritage goes back to 1979 when writing files to magnetic tape was a common method of storage.
Below is an example of using tar to compress and then extract files in an archive:
ls chimp.fasta human.fasta macaque.fasta orangutan.fasta # Pack into single file. The suffix is your responsibility. 'c' means create, and #'f' means that the next argument is the filename to write to. tar -cf sequences.tar *.fasta Note that the original files are untouched ls chimp.fasta human.fasta macaque.fasta orangutan.fasta sequences.tar # Delete all sequences rm *.fasta # 'x' means extract tar -xf sequences.tar ls chimp.fasta human.fasta macaque.fasta orangutan.fasta sequences.tar
Over time, the features of tar have increased to make it more convenient and modern versions are now capable of packing and compressing files, as in the example above.
ls chimp.fasta human.fasta macaque.fasta orangutan.fasta sequences.tar # Pack and gzip sequences simultaneously ( 'z' tells tar to use gzip) tar -zcf sequences.tgz *.fasta #List the contents without extracting tar -ztf sequences.tgz chimp.fasta human.fasta macaque.fasta orangutan.fasta # More recent versions of tar can also bzip2 files tar -jcf sequences.tbz2 *.fasta tar -jtf sequences.tbz2 chimp.fasta human.fasta macaque.fasta orangutan.fasta
Compression and decompression are actually done by the same program. Decompression program names like 'gunzip' are actually just convenient aliases that tell the computer to call the gzip program with the unzipping flags.
Working on remote computers
Why use a remote computer? There are many reasons: First, central computing resources tend to be much larger, more reliable and more powerful than your laptop or PC – if you need to do a lot of work or use a lot of data then you may have no option but to use a bigger computer.
There is also a world of difference between server-quality hardware and stuff on your desk. Uninterruptible power supplies, (i.e. backup batteries for when the power goes out) are one example. Servers also tend to have redundant components and memory that can detect and correct errors. At the top end, servers can detect and isolate faulty parts, report the problem, and continue running. Often the first time users of a central server know that a fault occurred is when an engineer turns up with a replacement part.
If you have a job that will take a long time to run, for instance Bayesian phylogenetic methods, you may not want to commit to leaving your personal computer untouched for long enough to complete the analysis (and you really trust your colleagues not to turn it off?) whereas central facilities are permanently on and have batteries to prevent small glitches in the power supply from affecting the computers. Lastly, and most importantly, central computers tend to have much more rigorous and tested policies for backing up data – Do you do regular backups? Are they kept in a separate physical location from the original? When was the last time you checked that the backup actually worked?
SSH (short for Secure SHell) is a method of connecting to other computers and giving access to a command-line on them; once we have a command-line we can interact with the remote computer just like we interact with the local one using the command-line. SSH replaces an older method of connecting to remote computers called telnet, which sends everything – including your password – as normal undisguised text so anyone can read it. It is not a good idea to use telnet unless you know what you are doing and you have no other option. Similarly, avoid FTP `File Transfer Protocol' for transferring files if you have sftp or scp available.
As well as keeping communications between your computer and a remote computer secure, SSH also allows you to verify that the remote computer is the computer it claims to be – no point keeping traffic secure if you send it to the wrong place – and prevents someone sitting in the middle of the connection listening to each message then passing it on, pretending to each side to be the other. (This is known as a Man-in-the-Middle attack http://en.wikipedia.org/wiki/Man-in-the-middle_attack. Both sides think they are communicating with the other but are actually communicating with an intermediary who copies all messages then forwards them on.) The method use to verify identity, without possibility of forgery, and even if someone else can copy and manipulate all messages is very interesting and has many other uses. See http://en.wikipedia.org/wiki/Public-key_cryptography and http://en.wikipedia.org/wiki/Digital_signature for details. If verification fails, you will be warned with a message like in Figure 13 and the computer will refuse to connect.
By far, the majority of these warnings are caused by inept computer administration rather than malice (for instance, if someone has upgraded the other machine incorrectly so it appears to be a different computer, you will get this kind of error). If you are sure it is safe, the warning can be dealt with by deleting the appropriate line for the computer from the ~/.ssh/known_hosts file. Graphical programs can also be run on remote machines, but expect pauses unless you have a very, very fast internet connection. The system that enables this is called the X Windows system (or just X, or X11) (X is the successor to the W Windows System, if you are wondering where the X came from). You can use the -X flag when you run ssh to allow the remote computer to programs in new windows on your local display, provided you have software on your local computer that understand the instructions being sent. Linux computers use such software by default for display and Mac OS X comes with software that can be used (and is started automatically by ssh in the following example). On Windows, the Cygwin software provides the required functionality. Below is an example of using ssh with the -X flag.
Transferring files
It is possible to transfer files between computers using SSH alone but this is not recommended since more friendly interfaces exist. Of course, there are many graphical file transfer programs available. Without recommending particular programs, Cyber-duck http://cyberduck.ch/ for the Mac OS X and WinSCP http://winscp.net/ for Windows appear to be usefule options, but there are many more. Alternatively, under Mac and Unix, it is possible to mount directories on remote computers so there appear to be local; search for sshfs for details. When transferring files, silent errors are extremely rare but can happen and so we'd like to be able to verify that the file received is identical to the one sent. Short files could be checked by eye, but this can't be automated without transferring the file again (which might also get an error). A common technique to verify correct transfer is to calculate the md5 (Message Digest algorithm 5) of both files and compare these values. The md5 is short string of characters that identifies a file and two different files are extremely unlikely to share the same string – if a file changes, its md5 will (very probably) change and so we know that that a change occurred. It is extremely difficult to deliberately create two files that have the same sum. The chances of two non-identical random files having the same md5 is about 3.4e38. When checking large numbers of files, the chance that there are two files in the set with the same md5 increases rapidly but will still be small enough for realistic uses. More rarely, you may come across SHA sums, shasum on both Unix and Mac computers, which are very similar to md5's but have an even smaller chance that two files share the same string.
Getting help
General help with Ubuntu has already been covered in “Acclimatisation“, alternatively, just find someone to ask. As with everything else, the web is a rich source of good, bad, and down-right weird tutorials.
If you are have little or no programming experience, Python (http://python.org/) is a good choice for learning how to do useful bioinformatics scripting, especially in conjunction with the Biopython module (http://biopython.org/). Unix is generally very well documented, although the documentation is often aimed at more experienced users. The manual pages all tend to follow the same format, and it's a good idea to become familiar with it. The page will start with a description of what the command does and a summary of all its flags. Optional flags will be enclosed in square brackets. Next comes a full description of the command and detailed descriptions of what each flag does. Sometimes there is also a section containing examples of usage. Mac OS X is generally very consistent about man pages but Linux derivatives can be a mixed bag.
Variables and programming
So far, we have only used the command-line to run other programs and to chain them together to achieve more complex results. The command-line tools can be used like a programming language in its own right, and we can write little programs to automate common tasks; often this referred to as scripting rather than programming although the distinction is not really relevant.
Obviously learning to program is not something that can be taught in an hour or two, and even experienced programmers take several days to become productive in a new language, so this section can give little more than a taste of what is possible; however, it should be possible to show how you could save a lot of time with a little investment up front. If you are doing similar things to large number of files, many sequences for example, typing the same command over and over on the command line is time-consuming, tedious, and prone to error, especially as you get bored. Scripting can save you a lot of time and allow you to get on with something else while the computer takes on that task for you. Think about the last time you needed to rename 100 files, or change the format of thousands of gene alignments so they are compatible with your phylogeny program. Learning a little bit of scripting can speed up these tasks tremendously. As with everything, there are many tutorials available on the web and a search for bash scripting tutorial or bash scripting introduction will yield many examples of varying completeness and comprehensibility.
In order to provide you with a little bit of programming background, we've prepared a small general tutorial below:
The first thing to introduce are variables. A variable is just a name for another piece of data, a useful analogy is that of a labelled box: every time we see the label, we replace it conceptually with the contents of the box. The ability to manipulate variables, changing the state of the computer, is fundamental to programming. Here we'll introduce two useful cases: shortening common directory paths and performing the same operations on many files. In bash scripting, variables called with a dollar sign, followed by the name of the variable: $NAME. There are some restrictions on the characters that can be part of a variable name, and variable names cannot start with a number. As a rule of thumb it's a good idea to only use upper- or lower-case letters in your variable names
A variable can refer to the name of a file and we can write things at the command-line using the variable instead of the name explicitly – change the variable and we run exactly the same commands on a different file. One way to take advantage of this this would be to set the variable to one of several files and use the history to repeat a set of commands. Of course, if the commands write their output to a file then that would have to be renamed each time otherwise the output for each file would be written over that for the previous. Shell scripting provides an alternative: the computer can be told to set the variable to each of many file names in turn and the value of the variable can be edited automatically to provide the name of a unique output file.
A common Unix practice is to place frequently used sets of functions into a file, called a script, for reuse thereby preventing errors retyping them. Writing a script also means that complex operations with many steps can be tested before you commit to running them over many files, something that could potentially take days if we are dealing with large numbers of genes. Scripts can be written and modified in any common text editor but must be saved in text format; nano is a good basic editor that is fairly intuitive to use but there are many others more specifically designed with programmers in mind. Alternatively you could use gedit, a program more like Notepad on Windows (to access gedit, click the Ubuntu button and search for gedit; entering gedit & at the command-line will also work).
Line endings – compatibility problems
Even after the standard alphabet for computers was established (ASCII – American Standard Code for Information Interchange) there was no agreement about how to how to indicated the end of a line. ASCII provides two possibilities: line-feed '\n' and carriage-return '\r' , based on how old type-writers and tele-type terminals used to work: a carriage-return moves the carriage, the position to print the next character at, back to be beginning of the line and line-feed moves the paper one line down but doesn't change where the carriage is. On Unix a '\n' character is taken to mean “line-feed and carriage return” and this is used to separate lines of text. On Windows, lines are separated by the pair of characters '\r\n' (in that order) and old versions of Apple operating systems (prior to OS X) use '\r' to separate lines. The situation on Mac OS X is more complex since it must deal with both its Mac and Unix heritage; officially '\n' ' now separates lines in files but programs have to be able to deal with both conventions.
To further complicate things, some methods of transferring files between machines try to automatically convert the line endings for you. This is generally a mistake. Specifically an old file transfer method called FTP “File Transfer Protocol” has two modes: text and binary, text mode will attempt to translate line endings. Unix platforms default to binary and are generally safe. The only case where you need to be careful is transferring files from Windows using the command-line FTP application. If you transfer a binary file over FTP in text mode, the received file will be corrupted irretrievably. If in doubt, see Transferring files for how to verify that your file has transferred correctly.
If you've managed to read through to here, you're probably thinking: a) that's complicated, and b)why haven't I noticed this? The answer is that it used to cause problems in the past but programmers are aware of the issues nowadays and programs tend to do the right thing. Some programming languages like Perl even deal with these problems transparently so even programmers don't need to be aware of them any more.
GUIs
Galaxy
Galaxy is an open source GUI toolset for a variety of NGS applications. It can be accessed through the Penn State University server[5] or an institution may set up their own server. Users can save and publish data histories for future use or for others to use. Its tutorials and graphic interface make it simple to learn and easy to use.
Users may also want to run their own galaxy instance, for testing, tool development or production use. A preconfigured Docker container makes this straightforward.
The following demonstrates an example setup.
The user:
- downloads and installs Docker (https://www.docker.com)
- pulls the galaxy-stable image from Docker Hub (https://hub.docker.com/)
docker pull bgruening/galaxy-stable
- runs the docker container
docker run -itp 8080:80 -v /local/dir/:/export bgruening/galaxy-stable
- can now access the fully functional galaxy instance at http://localhost:8080
GeneTalk
5 steps to analyze VCF files in GeneTalk at www.gene-talk.de
- Upload VCF file
- Edit pedigree and phenotype information for segregation filtering
- Filter your VCF file by editing the following filtering options
- Functional filter (filter out variants that have effects on protein level)
- Linkage filter (filter out variants that are on specified chromosomes)
- Gene panel filter (filter variants by genes or gene panles, subscribe to publically available gene panels or create own ones)
- Frequency filter (show only variants with a genotype frequency lower than specified)
- Inheritance filter (filter out variants by presumed mode of inheritance)
- Annotation filter (show only variants that are listed in databases)
- View results and annotations
- Write your own annotations
BaseSpace
References
- ↑ "GNU Operating System". Free Software Foundation, Inc. 11 April 2016. Retrieved 28 April 2016.
- ↑ "What is Copyleft?". Free Software Foundation, Inc. 3 October 2015. Retrieved 28 April 2016.
- ↑ Ubuntu Documentation Team. "Official Ubuntu Documentation". Canonical Ltd. Retrieved 28 April 2016.
- ↑ McIlroy, M.D.; Pinson, E.N.; Tague, B.A. (1978). "Unix Time-Sharing System: Forward". The Bell System Technical Journal. 57 (6): 1899–1904.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ "Galaxy". usegalaxy.org. Galaxy Project. Retrieved 28 April 2016.
Pre-processing
Pre-processing
FASTQ files – discussion of the various quality encodings
FASTQ files extend sequence information in FASTA files by also including information about sequence quality. A FASTQ file typically consists of four lines.
- A line starting with @ and containing the sequence identifier
- The actual sequence
- A line starting with + followed by an optional sequence identifier
- A line with quality values encoded in ASCII space
As such the 2nd and 4th line must have the same length One such entry is given below showing one sequence "ATGTCT"...
@HWI-ST999:102:D1N6AACXX:1:1101:1235:1936 1:N:0: ATGTCTCCTGGACCCCTCTGTGCCCAAGCTCCTCATGCATCCTCCTCAGCAACTTGTCCTGTAGCTGAGGCTCACTGACTACCAGCTGCAG + 1:DAADDDF<B<AGF=FGIEHCCD9DG=1E9?D>CF@HHG??B<GEBGHCG;;CDB8==C@@>>GII@@5?A?@B>CEDCFCC:;?CCCAC
Here a quality value ranging from -5 to 41 is added to an offset and the resulting character is taken from an ASCII table. As such the whole data can be represented as text. Whilst Illumina made multiple changes to the quality format and eventually returned to almost Sanger encoding, the most important difference is whether the offset is 33 as in Sanger and Illumina v1.8 and later or 64 as in previous Illumina (and Solexa) formats. As you can see from the chart if you find any of the following characters: !"#$%&'()*+,-./0123456789: your offset must be 33 whereas any of the following characters KLMNOPQRSTUVWXYZ[\]^_`abcdefgh point towards an offset of 64. The above example is thus base offset by 33 as we find a 1 as first character. Also bear in mind that the @ and + signs are valid characters for quality so even if a line started by @ or + this could just be the beginning of the quality string.
See the quality chart below which is modified from the wikipedia article.
SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS..................................................... ..........................XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX...................... ...............................IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII...................... .................................JJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJ...................... LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL.................................................... !"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHIJKLMNOPQRSTUVWXYZ[\]^_`abcdefghijklmnopqrstuvwxyz{|}~ | | | | | | 33 59 64 73 104 126 0........................26...31.......40 -5....0........9.............................40 0........9.............................40 3.....9.............................40 0........................26...31........41
S - Sanger and Illumina 1.8+, Offset 33, Quality range (0, 40) (!,I) X - Solexa, Offset 64, Quality range (-5, 40) (;,h) I - Illumina 1.3+ Offset 64, Quality range (0, 40) (@,h) J - Illumina 1.5+ Offset 64, Quality range (3, 40) (B,h) L - Illumina 1.8+ Offset 33, Quality range (0, 41) (!,J)
Presentation of the metrics used in QC
When you turn to quality control there are various metrics to consider.
Sequence Quality
The simplest is obiviously the quality score introduced in the FASTQ files above. As such it gives already a valid idea about base call quality. As often quality of reads degrades over the course of a sequence it is common practice to determine the average quality of the first, second, third,...nth base by just averaging over all reads in a file. Also to give some idea about the spread usually bar plots showing quantiles are given. This would give us an idea about what kind of trimming and quality filtering the data requires.
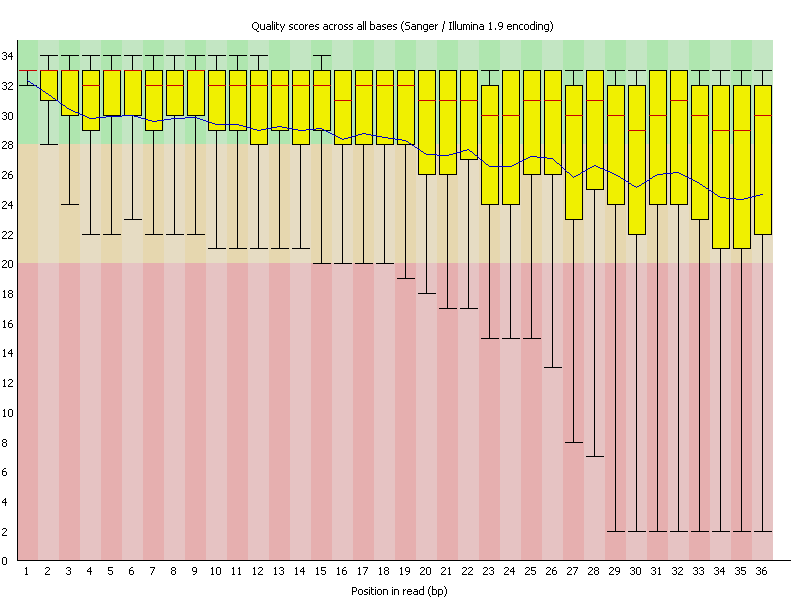
As an example here sequence data was investigated for quality using FastQC. As you can see the sequence reads are 36 bases long and the average sequence quality (depicted by the blue line) is steadily declining. In many new Illumina kits the sequence quality goes up a bit first before it steadily declines.
However instead of going over each base one can average the quality of each read instead and show a cumulative plot of the sequence quality of these.
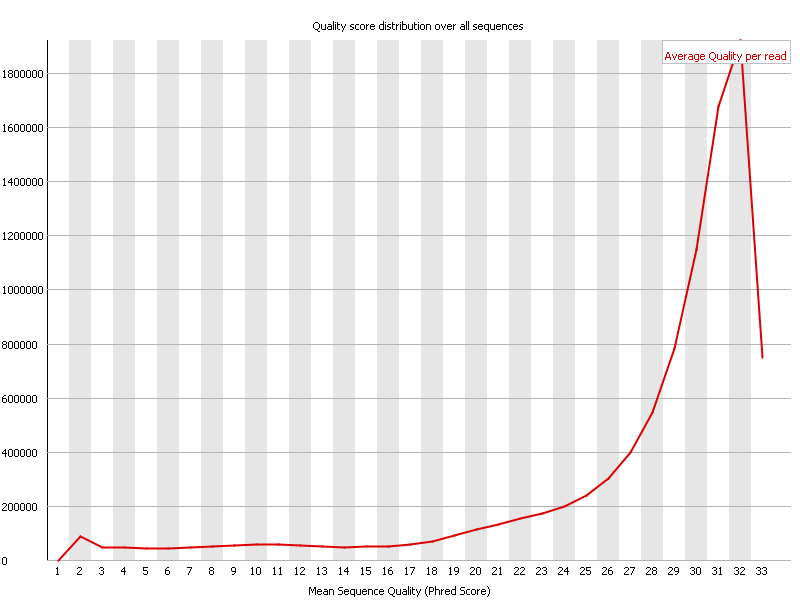
In the above screenshot one can observe that most reads have an average quality of 32. This is to be considered very good in general, however given that these reads are somewhat on the short side, it is probably at best an OK result.
Per Base Sequence Content
Another important metric is to look for base content at each position. Assuming the data is a random sample from the sequence space, at each position the contribution should be identical. Thus one needed to see straight lines. In reality it often happens that the first few bases might indeed show some erratic behavior, which could be due to non completely random primers. In the shown example however the reads are completely off. As you can see there is considerable bias in each base over the whole reads. In fact this bias is so strong, that you can read the overrepresented bases of the read.

As an example if you look at the last few bases you can read them as CTTGAAA-end of sequence.
Adapter sequence present or not?
If we now turn our attention to the overrepresented sequences in FastQC we can immediately figure out where this came from:
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCTCGTATGCCGTCTTCTGCTTGAAA | 1870684 | 19.446980406066654 | Illumina Single End Adapter 1 (100% over 33bp) |
| GAAGAGCTCGTATGCCGTCTTCTGCTTGAAAAAAAA | 95290 | 0.9906017065918623 | Illumina Single End Adapter 1 (100% over 28bp) |
Intro to errors and quality scores/encoding
As mentioned above the base caller assigns a quality score which is then available for each base. This gives the estimated reliability for this base. Please note that depending on your sequencing platform the typical mistakes are different. Illumina's most prevalent form of mistake is a nucleotide exchange whereas 454, Ion Torrent/Proton and other similar platforms have major issues with homopolymers such as AAAAAA where the correct number of As can often not be determined exactly.
Preprocessing Steps
Sequence Quality Trimming
In order to cope with lower quality data it is common to remove low quality bases. Typically one would remove lower quality bases from the e.g. the 3' end using a sliding windows approach as the per base quality gradually drops.
Alternative clipping strategies (Adaptor clipping)
In addition to removing lower base quality data, one would also remove adapters, PCR primers and other artifacts. In practice one would combine the adapter clipping with quality trimming approaches.
K-mer filtering/correction strategies
There are different ways to correct for base errors using kmer approaches because some errors can not be simply clipped off and even a very good quality value does not mean that a read is really error free. This is an important step prior to an assembly but potentially less crucial for alignments.
One basic idea is based on kmers in the read string. The original idea going back at least until 2001 (Pevzner 2001) generates a spectrum of kmers first, then kmers which are above a certain threshold (called solid) and kmers below this threshold are potentially arising from mistakes.
If a read is split into multiple kmers a single sequencing error will result in converting several overlapping kmers from strong to weak ones. An error correction step could now try to find the smallest number of changes required to make all kmers in the read strong.
Variants such as Quake also take the base quality into account to be better able to discriminate between low copy true kmers and high copy error kmers.
Digital Normalization and Partitioning
When considering especially RNA sequencing, it is well known that a normalization of RNA using molecular biology techniques (lab normalization) can help in providing better contigs and that a better general representation is achieved. This is because using lab normalization depletes common -or highly represented- sequences. Thus, when sampling from sequence space, after lab normalization it is less likely to find the previously very common sequences and thus more likely to find the previously underrepresented sequences. Apart from the advantage of having a higher likelihood to find underrepresented sequences there is the additional advantage that it is now less likely to find the same sequencing error multiple times in two or or more independent reads due to sheer oversampling. The latter makes it less likely that assembly software would erroneously create multiple contigs out of one true mRNA, due to these correlated SNPs. That said, lab normalization is neither easy and if it is outsourced it can be costly. Thus, one can instead use digital normalization. The basic idea is to downsample reads that have a lot of abundant kmwers. In addition this has the added benefit (to the ones above) that the number of reads to process becomes smaller, and thus it might be much more feasible (and faster) to assemble a transcriptome. One way to go about this is to use Titus Brown's tool set: http://ged.msu.edu/papers/2012-diginorm/
Paired end merging
A number of tools will take Illumina paired-end data and merge the reads if an overlap can be detected between them, potentially correcting errors by a taking the higher quality basecall at discrepant positions. This may improve assembler performance by reducing the data complexity, and may also improve the resulting contigs by removing erroneous data and improving the assembly of repeats. Tools to accomplish this include COPE and FLASH.
Removal of other undesirable sequences
Depending on the design of an experiment, there may be other sequences which are desirable to remove or mask from the reads prior to assembly. For example, if sequencing pools of BAC or cosmid DNA, it may be desired to remove most if not all of the vector backbone. Similarly, E.coli sequences will contaminate BAC or cosmid DNA preparations and could be removed in advance. Removing these post-assembly is an option as well. The PhiX control viral DNA is a common contaminant in Illumina sequencing data. Fast search tools such as SMALT can be used to map reads against a reference genome in order to identify those which should be removed.
Exercise
QC workflow (Data from machine -> pre-trimming quality plots -> trimming -> post-trimming quality plots)
A typical workflow might thus be to first get the data from the machine, and evaluate the typical quality plots as shown in the previous section. This gives a valid and important insight into the read quality and might potentially raise awareness about library preparation problems that might have occurred. After this problems have been identified and noted down, one would try to remove several errors by using trimming tools such as Trimmomatic to remove low quality bases from the sequence end and (potentially more importantly) to also remove remaining adapters etc from the reads. After having thus processed the reads, one would once again judge the quality to inform about remaining quality issues. As an example, even after removing known adapters from the sequences as in the above case, one might still see a per base sequence bias and would want to remove this bias or at least keep it in mind. We will discuss one exemplary workflow here.
The dataset
Download SRR074262 from the SRA (this is the example from above).
Fastq output analysis
Download FastQC (or analyze your data in RobiNA for similar plots). FastQC is relatively self explanatory. Open the FastQ file you just downloaded. FastQC will run through your dataset and you generate the plots shown in the introduction by clicking on the individual categories on the left hand side.
Adapter removal only
We will use Trimmomatic to simply remove adapters. java -jar trimmomatic-0.30.jar SE -phred33 SRR074262.fastq aclipped.fq.gz ILLUMINACLIP:TruSeq2-SE.fa:2:30:10 MINLEN:25 This tells trimmomatic that the quality encoding is phred 33 (modern Illumina) and it will store the results in the compressed file adapter_clipped.fq.gz. Finally it will use TruSeq3 adapters provided by trimmomatic.
Trimmomatic report
Input Reads: 9619406 Surviving: 7443332 (77.38%) Dropped: 2176074 (22.62%) TrimmomaticSE: Completed successfully
FastQC automatically recognizes gz files, so having the file compressed is ok. Indeed when we open the file aclipped.fq.gz in FASTQC the adapters have been removed.
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GGGGAGGAGAGAGCCATTGTTGAGGCGGCCATTGAG | 46910 | 0.630228505190954 | No Hit |
| AGGGGAGGAGAGAGCCATTGTTGAGGCGGCCATTGA | 21029 | 0.28252132244000405 | No Hit |
Adapter Removal and quality clipping
We will use Trimmomatic to remove adapters and use a sliding window approach to remove lower quality bases at the end
java -jar trimmomatic-0.30.jar SE -phred33 SRR074262.fastq aclippedtrim.fq.gz ILLUMINACLIP:TruSeq2-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:25
Input Reads: 9619406 Surviving: 6513310 (67.71%) Dropped: 3106096 (32.29%) TrimmomaticSE: Completed successfully
Read correction
Links to useful tools for preprocessing
Links to resources used in this chapter
Links to other QC tools e.g. FASTX toolkit and spectral alignment for error correction
Alignment
This page in a nutshell: Given sequencing data (reads) and the reference sequence for the species, comparing the reads to the reference is an easy way to detect small variations in the sequenced sample, such as SNPs and short InDels. |
Introduction
Alignment, also called mapping,[1] of reads is an essential step in re-sequencing. Having sequenced an organism of a species before, and having constructed a reference sequence, re-sequencing more organisms of the same species allows us to see the genetic differences to the reference sequence, and, by extension, to each other. Alignments of data from these re-sequenced organisms is a relatively simple method of detecting variation in samples. There are certain instances (such as new genes in the sequenced sample that are not found in the existing reference sequence) that can not be detected by alignment alone; however, while other approaches, such as de novo assembly, are potentially more powerful, they are also much harder or, for some organisms, impossible to achieve with current sequencing methods.
Next-generation sequencing generally produces short reads or short read pairs, meaning short sequences of <~200 bases (as compared to long reads by Sanger sequencing, which cover ~1000 bases). To compare the DNA of the sequenced sample to its reference sequence, we need to find the corresponding part of that sequence for each read in our sequencing data. This is called aligning or mapping the reads against the reference sequence. Once this is done, we can look for variation (e.g. SNPs) within the sample. This poses a number of problems:
- The short reads do not come with position information, that is, we do not know what part of the genome they came from; we need to use the sequence of the read itself to find the corresponding region in the reference sequence.
- The reference sequence can be quite long (~3 billion bases for human), making it a daunting task to find a matching region.
- Since our reads are short, there may be several, equally likely places in the reference sequence from which they could have been read. This is especially true for repetitive regions.
- If we were only looking for perfect matches to the reference, we would never see any variation. Therefore, we need to allow some mismatches and small structural variation (InDels) in our reads.
- Any sequencing technology produces errors. Similar to the "real" variation, we need to tolerate a low level of sequencing errors in our reads, and separate them from the "real" variation later.
- We need to do that for each of the millions of reads in our sequencing data.
Short reads
Raw short reads often come in (or can be converted into) a file format called FASTQ.[2] It is a plain text format, containing the sequence and quality scores for every read, where each single read normally occupies four consecutive lines:
@Read_id_1 CTGATGTGCCGCCTCACTTCGGTGGT + @@@DDDDDH8<BAHG@BHGIHIII>( @Read_id_2 TGATGTGCCGCCTCACTACGGTGGTG + FHHHHHJIJIJIJIIIJJIIJGIGII @Read_id_3 ...
The four lines are:
- The name/ID of the read, preceded by a "@". For read pairs, there will be two entries with that name, either in the same or a second FASTQ file.
- The sequence of the read.
- A "+" sign. In very old FASTQ files, this is followed by the read name from the first line. Today, this line is present for
historical reasonsbackwards compatibility only. - The quality scores of the bases from line 2. The scores are generated by the sequencing machine, and encoded as ASCII (33+score) characters. The line should have the same length as line 2, as there is one quality score per base.
Alignment
For each of the short reads in the FASTQ file, a corresponding location in the reference sequence (or that no such region exists) needs to be determined. This is achieved by comparing the sequence of the read to that of the reference sequence. A mapping algorithm will try to locate a (hopefully unique) location in the reference sequence that matches the read, while tolerating a certain amount of mismatch to allow subsequence variation detection. Reads aligned (mapped) to a reference sequence will look like this:
GCTGATGTGCCGCCTCACTTCGGTGGTGAGGTG Reference sequence CTGATGTGCCGCCTCACTTCGGTGGT Short read 1 TGATGTGCCGCCTCACTACGGTGGTG Short read 2 GATGTGCCGCCTCACTTCGGTGGTGA Short read 3 GCTGATGTGCCGCCTCACTACGGTG Short read 4 GCTGATGTGCCGCCTCACTACGGTG Short read 5
You can see the reference sequence on the top row, and five short reads stacked below; this is called a pileup. While two of the reads are a perfect match to the reference, the three other reads show a mismatch each, highlighted in red ("A" in the read, instead of "T" in the reference). Since there are multiple reads showing the mismatch, at the same position, with the same difference, one could conclude that it is an actual genetic difference (point mutation or SNP), rather than a sequencing error or mismapping.
Mapping algorithms
There are several alignment algorithms in existence; you can find an (incomplete) list further down in software packages. Some notes on mapping algorithms:
- The reference sequence, the short reads, or both, are often pre-processed into an indexed form for rapid searching. (See BWT)
Sources of errors
There are several potential sources for errors in an alignment, including (but not limited to):
- Polymerase Chain Reaction artifacts (PCR artifacts). Many NGS methods involve one or multiple PCR steps. PCR errors will show as mismatches in the alignment, and especially errors in early PCR rounds will show up in multiple reads, falsely suggesting genetic variation in the sample. A related error would be PCR duplicates, where the same read pair occurs multiple times, skewing coverage calculations in the alignment.
- Sequencing errors. The sequencing machine can make an erroneous call, either for physical reasons (e.g. oil on an Illumina slide), or due to properties of the sequenced DNA (e.g., homopolymers). As sequencing errors are often random, they can be filtered out as singleton reads during variant calling.
- Mapping errors. The mapping algorithm can map a read to the wrong location in the reference. This often happens around repeats or other low-complexity regions.
Alignment types
Alignments can be used for different purposes:
- Whole-genome sequencing. This would be the "default" use; sequence all DNA from an organism and map it to the appropriate reference sequence, to find genetic variation.
- Exome sequencing. For large genomes (e.g., human), capture just the exomic DNA before sequencing. This will return sequencing data for most of the genes, at a fraction of the cost.
- Transcriptome sequencing (RNA-Seq). Sequencing of the transcriptome, that is, of the RNA present in the sample. This can show which genes are transcribed in the sample, and help fine-tune gene annotation (exon boundaries etc.). Mapping can be done either to the full reference sequence, or to a special "transcriptome reference".
- ChIP-Seq (Protein-DNA interaction).
The SAM/BAM format
The SAM/BAM format has emerged as the de facto standard format for short read alignments. SAM[3] is the plain-text version of the binary, compressed BAM format. They can be converted into one another by the name-giving samtools[4] command-line tool. BAM (without alignment position data) is increasingly used as a space-saving alternative to FASTQ files for containing the short raw read data, and all current alignment software can generate SAM/BAM as an output format. Once in BAM format, the file can be indexed, giving quick access to any region of the reference sequence. Subsequently, using samtools or other software, BAM files can be analysed (e.g. for quality control), modified (removal of PCR duplicates, local realignment, base quality recomputation), or used to call variation, either small (SNPs, short InDels) or large (inversions, tandem duplications, deletions, translocations). BAM files can be visualised using tools like Artemis, ACT, or LookSeq[5]. Last but not least, alignments in BAM format can be used to "morph" the reference sequence to correspond to the short read data with ICORN[6]; this can be useful to get an actual DNA sequence for a sample, or to construct a new reference sequence based on a closely related species.
Other useful SAM/BAM-related software includes:
SAM/BAM tools examples
- Convert SAM to BAM format:
- samtools view -bS aln.sam > aln.bam
- Sort BAM file according to position on reference sequence
- samtools sort aln.bam aln_sorted.bam
- Index BAM file (needed for visualising the alignment)
- samtools index aln_sorted.bam aln_sorted.bai
- Extract the header information from a BAM file
- samtools view –h aln_sorted.bam > aln.sam
- Generate a FASTQ file from a BAM file
- bam2fastq -o aligned.fastq --no-unaligned aln.bam
Software packages
| Software | Type | Supported technologies |
Interface | Notes |
|---|---|---|---|---|
| Partek | Commercial | All | GUI |
|
| BWA[9] | Free software | Illumina SOLiD 454 |
Command line |
|
| Bowtie[10] | Free software | Illumina SOLiD |
Command line |
|
| Stampy[11] | Free software | Illumina | Command line |
|
| SHRiMP2 | Free software | Illumina | Command line |
|
| TMAP | Free software | IonTorrent | Command line |
|
| SNP-o-matic[12] | Free software | Illumina | Command line |
|
| CLC workstation | Commercial | All | GUI |
|
| NextGenMap[13] | Open source | Illumina, Ion Torrent | Command Line | Fast and accurate. Self adjusting to the underlying data. Robust for high polymorphism
|
| Novoalign | Commercial for multi-threaded version. Single threaded version is free | Illumina | Command Line | Fast and accurate. Probably the best aligner as of 2013. |
| GSMapper | Commercial | 454 | GUI | / |
| SSAHA2 | Free software | 454 | Command line | Fast and accurate for all reads it can map |
| BLAT | Free software | 454 | Command line | Not designed for NGS data. |
| Mosaik | Free software | All | Command line | Tedious steps. Alignment speed can be slow. Huge memory requirement. |
| BWA-SW[14] | Free software | 454, IonTorrent | Command line |
|
| BWA-MEM[15] | Free software | 454, IonTorrent | Command line |
|
| Bfast[17] | Free software | SOLiD | Command Line | Speed of alignment may be too slow for large NGS data [18] |
| Tophat[19] | Free software | Illumina | Command Line | Transcriptome data only |
| Splicemap | Free software | Illumina | Command Line | Transcriptome data only |
| MapSplice | Free software | Illumina | Command Line | Transcriptome data only |
| AbMapper | Free software | Illumina | Command Line | Transcriptome data only |
| ERNE-map (rNA)[20] | Free software | Illumina | Command line |
|
| mrsFAST-Ultra[22] | Free software | Illumina | Command line / GUI |
|
An exhaustive list of NGS aligner is maintained at HTS mapper
Based on personal experience and prevalence and based on literature data on the performance[23][24][25], a quick primer on when to use which software:
- If only speed matters use bowtie.
- BWA is a bit slower but more sensitive.
- If sensitivity and specificity is needed, try NextGenMap, Stampy, Novoalign, or SHRiMP 2.
Furthermore a framework (Teaser) was proposed that automatically benchmarks several mappers within minutes.[26]
References
- ↑ Flicek, P.; Birney, E. (2009). "Sense from sequence reads: Methods for alignment and assembly". Nature Methods. 6 (11 Suppl): S6–S12. doi:10.1038/nmeth.1376. PMID 19844229.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. (2010). "The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants". Nucleic Acids Research. 38 (6): 1767–71. doi:10.1093/nar/gkp1137. PMC 2847217. PMID 20015970.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ The SAM/BAM Format Specification Working Group (18 November 2015). "Sequence Alignment/Map Format Specification" (PDF). GitHub. p. 16. Retrieved 28 April 2016.
- ↑ "Samtools organisation and repositories". GitHub. Retrieved 28 April 2016.
- ↑ Manske, H.M.; Kwiatkowski, D.P. (2009). "LookSeq: A browser-based viewer for deep sequencing data". Genome Research. 19 (11): 2125–2132. doi:10.1101/gr.093443.109. PMC 2775587. PMID 19679872.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ "ICORN: Iterative Correction of Reference Nucleotides". SourceForge. Retrieved 28 April 2016.
- ↑ Broad Institute. "Picard". GitHub. Retrieved 28 April 2016.
- ↑ Stein, L.D. (12 February 2016). "Bio-SamTools". CPAN. Perl.org. Retrieved 28 April 2016.
- ↑ Li, H.; Durbin, R. (2009). "Fast and accurate short read alignment with Burrows–Wheeler transform". Bioinformatics. 25 (14): 1754–1760. doi:10.1093/bioinformatics/btp324. PMC 2705234. PMID 19451168.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. (2009). "Ultrafast and memory-efficient alignment of short DNA sequences to the human genome". Genome Biology. 10 (3): R25. doi:10.1186/gb-2009-10-3-r25. PMC 2690996. PMID 19261174.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Lunter, G.; Goodson, M. (2011). "Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads". Genome Research. 21 (6): 936–939. doi:10.1101/gr.111120.110. PMC 3106326. PMID 20980556.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Manske, H.M.; Kwiatkowski, D.P. (2009). "SNP-o-matic". Bioinformatics. 25 (18): 2434–2435. doi:10.1093/bioinformatics/btp403. PMC 2735664. PMID 19574284.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Cibiv. "NextGenMap". GitHub. Retrieved 28 April 2016.
- ↑ Li, H.; Durbin, R. (2010). "Fast and accurate long-read alignment with Burrows–Wheeler transform". Bioinformatics. 26 (5): 589–595. doi:10.1093/bioinformatics/btp698. PMC 2828108. PMID 20080505.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Li, H. (2013). "Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM". Preprint: 1–3.
- ↑ Chapman, B. (6 May 2013). "Framework for evaluating variant detection methods: Comparison of aligners and callers". Blue Collar Bioinformatics. Retrieved 28 April 2016.
- ↑ Homer, N. "Blat-like Fast Accurate Search Tool". SourceForge. Retrieved 28 April 2016.
- ↑ Koboldt, D. (3 July 2011). "Aligners". MassGenomics. Retrieved 28 April 2016.
- ↑ Kim, D.; Salzberg, S. (23 Februart 2016). "TopHat". Johns Hopkins University Center for Computational Biology. Retrieved 28 April 2016.
{{cite web}}: Check date values in:|date=(help)CS1 maint: multiple names: authors list (link) - ↑ Vezzi, F.; Del Fabbro, C.; Tomescu, A.I.; Policriti, A. (2011). "rNA: a fast and accurate short reads numerical aligner". Bioinformatics. 28 (1): 123–124. doi:10.1093/bioinformatics/btr617. PMID 22084252.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ "Welcome to ERNE 2!". SourceForge. 29 September 2014. Retrieved 28 April 2016.
- ↑ Hach, F.; Sarrafi, I.; Hormozdiari, F.; Alkan, C.; Eichler, E.E.; Sahinalp, S.C. (2014). "mrsFAST-Ultra: A compact, SNP-aware mapper for high performance sequencing applications". Nucleic Acids Research. 42 (W1): W494–W500. doi:10.1093/nar/gku370. PMC 4086126. PMID 24810850.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Bao, S.; Jiang, R.; Kwan, W.; Wang, B.; Ma, X.; Song, Y.Q. (2011). "Evaluation of next-generation sequencing software in mapping and assembly". Journal of Human Genetics. 56 (6): 406–14. doi:10.1038/jhg.2011.43. PMID 21525877.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Li, H. (19 November 2009). "NGS Alignment Programs". SourceForge. Retrieved 28 April 2016.
- ↑ Li, H. (19 November 2009). "NGS mapper ROC curves". SourceForge. Retrieved 28 April 2016.
DNA Variants
DNA Variants
Protocols
Whole genome, exome, etc. Consequences for downstream analysis
Typical workflow
File formats
VCF
VCF stands for Variant Call Format. It was created by the 1000 Genomes Project as a way to store small-scale variation data (SNPs, InDels, short structural rearrangements), and has since become the de facto standard format for storing such data. The official, detailed description can be found here (VCF version 4.1, as of writing).
VCF can store information about a variant, such as its position on a reference sequence, the reference and alternate alleles, stable variant identifier (e.g. rs number), as well as the observed allele(s) in multiple samples. VCF can also hold aggregate information about the variant across all samples (e.g. total coverage depth, allele frequencies etc.), as well as a list of filters that the variant failed during the current analysis.
The basic VCF file format is ASCII text. A header section identifies the VCF format version, defines FILTER and INFO fields, and other meta-data. This is followed by the actual data table, consisting of a single row containing the standard headers and the sample names, and one row per variant. All columns in the table header and the data rows are separated by tab (\t) characters:
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT Sample1 Sample2 Sample3 2 4370 rs6057 G A 29 . NS=2;DP=13;AF=0.5;DB;H2 GT:GQ:DP:HQ 0|0:48:1:52,51 1|0:48:8:51,51 1/1:43:5:.,.
(for more exhaustive examples, see the official description)
Creating a dataset
SAMTOOLS
SAMtools is a library and software package that manipulates alignments in SAM/BAM format. The format of the alignments are human readable. This software helps to convert from other alignment formats. It also can sort and merge the alignments. PCR duplicates also can be removed using SAMtools.[1]
SAMtools has two separate implementations one in C and in Java which are slightly different in function. The implementation comes as a library in C and a command line tool that packages several utilities including[1]:
- import: SAM-to-BAM conversion
- view: BAM-to-SAM conversion and subalignment retrieval
- sort: sorting alignment
- merge: merging multiple sorted alignments
- index: indexing sorted alignment
- faidx: FASTA indexing and subsequence retrieval
- tview: text alignment viewer
- pileup: generating position-based output and consensus/indel calling
others...
Reference datasets
Human=> Variants=>1000 genomes, HapMap,etc
Other species
Viewing datasets
Ensembl
UCSC
IGV
Tablet?
Comparing datasets
VCF tools
SEQwiki content dump
SNP detection
SNPs, or single nucleotide polymorphisms, are heritable single base changes in a genome versus a reference sequence. They are part of the more generic set of Single Nucleotide Variations (SNVs), which also encompasses somatic single base changes which are not passed to offspring and are due to environmental damage. Tools for SNP identification can also be used for SNV identification, though tools specific for SNV identification exist as well. In some contexts, such as cancer genomes, SNV identification is complicated by heterogeneous DNA samples.
SNP identification programs must distinguish system noise (instrument errors, PCR errors, etc) from actual variation. They generally do so by modeling various error types and the expected distribution of calls under homozygous reference (AA), homozygous variant (BB) and heterozygous variant (AB) states. Confidence in calls is generally affected by the reported sequence quality values and read depth. Some SNP/SNV callers work by comparing individual samples to a reference, whereas others can simultaneously call in multiple samples using information from each sample to assist calling in the other samples. SNP callers for mixed population samples also exist.
A common source of error in SNP/SNV calling is misalignment due to pseudogenes, repeated genomic segments or close orthologs; in these cases the co-alignment of reads arising from different genomic regions can result in a false positive call. Another source of error can be local misalignment (or ambiguous alignment) due to indels in reads (either true indel variations or sequencing errors); realignment tools such as Dindel and those found in GATK can generate more consistent treatment of indels to reduce this source of error. Many SNP/SNV callers are designed for diploid DNA, and may not work well in samples with higher ploidy. As noted above, heterogeneity in samples such as tumor samples can frustrate SNV calling, and some callers are specifically designed to cope with this. Tumor samples may also have altered copy number due to gene or chromosomal amplification, meaning they are effectively of triploid or higher ploidy in some regions.
SNP/SNV callers often call only these polymorphisms, and not (for example) small indels. Users of these tools should also take care when calling adjacent pairs of SNPs/SNVs, as the phasing of these (or more distant SNPs) is not reported in many callers' reports.
Decision Helper
I want to quickly call SNP versus a reference =>Freebayes, samtools
Software Packages
Free Software
Freebayes
Freebayes[2] is the successor of Poly- Giga- and BAMBayes and should be much faster than these. Like these it relies on BAM files. It has also been described in some more detail by its developer on Biostar.[3]
- Pros
- very easy to run for simple SNP calling
- Does not assume any ploidy
- can read BAM files via STDIN
GATK
The Genome Analysis toolkit GATK allows multiple steps. The authors used their pipeline for variant calling using the NA12878 exome data set and compared their results to those of Crossbow (which uses SOAPsnp). Based on these results they concluded that crossbow had a lower specificity.
One easy way to to run GATK and other tools might be to use the variant pipeline Pipette mentioned on Biostar.[4][5]
- Important reminder
- If you run GATK framework in your own pipeline, you have to bear in mind GATK has Stringent file formatting requirement.
- e.g. chromosomes ordering in genome reference file has to be in canonical order.[6]
- BAM header has to be present in every BAM file.
- The BAM file has to be sorted, preferably by Picards because it write the proper header after sorting
- Read-group tag has to be present in each BAM. Either input the correct tag during mapping or you may waste your time in fixing the BAM file afterwards
- Pro
- Likely relatively specific (The authors show higher specificity than crossbow)
- Con
- relatively complex pipelines
MAQ
MAQ
- Pros
- performed slightly better than soapsnp and better than snvnmix according to an independent comparison
samtools
samtools using the mpileup command[7]
samtools pileup (without the m) is deprecated and has been removed in recent SAMtools versions.
Sibelia
Sibelia is a comparative genomic tool to assist biologists in analysing genomic variations that correlate with pathogens, or the genomic changes that help microorganisms adapt in different environments. Sibelia is also useful in evolutionary and genome rearrangement studies for multiple strains of microorganisms.[8]
- Pros
- Works well for multiple bacterial genomes.
- Easy to run and cross-platform, licensed under GPL.
- Cons
- Works slow for large genomes.
SOAPsnp
SOAPsnp is e.g. used in the Crossbow pipeline.
SNVMix
SNVMix The authors of SNVMix compared their tool to MAQ v0.6.8 and found better performance as judged by area under the curve when using Affymetrix SNP 6.0 data. However in an independent comparison using MAQ 0.71 MAQ performed better.
- Cons
- Might be unstable in high coverage region according to an independent comparison.
- Might be less precise than MAQ and SOAPsnp
VariationHunter
VariationHunter-CommonLaw is a tool for discovery of structural variation using high throughput technologies.
- Pros
- Allows structural variation detection in one or more individuals simultaneously
deStruct
deStruct is a software tool for identifying structural variation in tumour genomes from whole genome Illumina sequencing.
- Pros
- High sensitivity and specificity
- Able to automatically distribute parallel jobs on Linux clusters (such as SGE)
- Low memory requirement
Commercial Software
Strand NGS
Avadis NGS
Partek
CLCBio
Further reading
- Further Reading
- Alkan et al., 2011 Finding variation using different approaches
- Original Publications
- Li et al., 2008 MAQ
- Li et al., 2009 SOAP2 (snp)
- Li et al., 2011 samtools (snp)
- Goya et al.,2010 SNVMIX
- DePristo et al., 2011 GATK (snp)
- Comparisons
- Nielsen R, Paul JS, Albrechtsen A, Song YS Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. (2011) 12:443-51. The article gives general reccommendations for a workflow and suggests to use a calibration step as implemented by GATK or SOAPsnp
- Wang et al., 2011 A comparison of short read aligners and performance assesment of MAQ (0.71), SOAPsnp (1.03) and SNVmix(2-0.11.8-r4) where MAQ performed best
References
- ↑ a b Li, H.; Handsaker, B.; Wysoker, A.; et al. (2009). "The Sequence alignment/map (SAM) format and SAMtools". Bioinformatics. 25 (16): 2078–9. doi:10.1093/bioinformatics/btp352. PMC 2723002. PMID 19505943.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ "ekg/freebayes". GitHub. 19 April 2016. Retrieved 30 April 2016.
- ↑ Lindenbaum, P. (8 April 2010). "Question: What Methods Do You Use For In/Del/Snp Calling?". Biostars. Biostar Genomics, LLC. Retrieved 30 April 2016.
- ↑ "metalhelix/pipette". GitHub. 27 June 2013. Retrieved 30 April 2016.
- ↑ Matthieu (12 May 2011). "Question: Workflow Or Tutorial For Snp Calling?". Biostars. Biostar Genomics, LLC. Retrieved 30 April 2016.
- ↑ Van der Auwera, G. (25 July 2012). "What input files does the GATK accept / require?". GATK Forums. Broad Institute. Retrieved 30 April 2016.
- ↑ "Calling SNPs/INDELs with SAMtools/BCFtools". GitHub. 17 December 2010. Retrieved 30 April 2016.
- ↑ "Sibelia (aka Synteny Block ExpLoration tool)". St. Petersburg Academic University of the Russian Academy of Sciences. Retrieved 30 April 2016.
RNA
This guide is meant to offer an easy to follow guide to the analysis of RNA-seq data, aimed at those without any prior experience analysing next-gen data. However, a basic level of familiarity with R, the next-gen sequencing procedures and using the UNIX shell are assumed. Most of the steps described here are outlined in the review article.[1]
- It was primarily written by Matthew Young (myoung@wehi.edu.au) and is a work in progress.
- The pathogen example was provided by B. Usadel and makes use of a different set of tools.
RNA
Introduction to transcriptomics..
For every sample on which RNA-seq is run, the output you will typically receive is a file containing millions of short (25-300bp) DNA sequences, called reads, and quality scores indicating the confidence of each base call. However, there are some important common variations on this which depend on the platform and protocol used. These include, but are not limited to:
- Base space or colour Space
- Paired end/mate pair or single end/unpaired
- Stranded or un-stranded
Each of these is described in more detail in the following sections.
Base space vs Colour space
The two main platforms for second-generation sequencing of RNA are produced by Illumina and ABI Solid. While both produce millions of short reads, they are sequenced and reported in slightly different ways. The Solid platform uses a sequencing technique which generates the read information by attaching two base pairs at a time. Each base pair of a read is sequenced twice, by two (potentially different) di-nucleotides. To take advantage of this sequencing chemistry, Solid reports its reads not as a sequence of nucleotides, but as a sequence of four colours, where each colour represents a transition between bases, known as "colour space encoding."[2] On the other hand, the Illumina platform reads one base at a time along a fragment until the desired read length is reached. The observed bases are then reported as the output.
The consequence of this is that the tools for analyzing RNA-seq data depend on the platform used to produce the short read data. While it is possible to convert colour space reads to base space (and vice versa), doing so introduces severe biases into the data which should be avoided at all costs. Therefore, reads should be kept in their native format and the appropriate tools should be used to analyze them.
Paired end/Mate pair reads
The standard RNA-seq protocol involves random shearing of reverse transcribed mRNA (cDNA), followed by sequencing a short "read" from one end of the fragment. This means that only the first 25-300bp of a fragment are known (depending on the length of the reads) with the rest of the fragment remaining unsequenced. In fact, because fragmentation is random, the length of each fragment is also unknown, although a size selection step is usually applied.
Although the chemistry is not sufficiently precise to allow the entire fragment to be sequenced, a clever trick can be applied whereby a short read is taken from both ends of the fragment resulting in a pair of short reads one from each end of the fragment. Reads where this has been performed are known as paired end or mate pair reads. Paired end reads allow additional information to be inferred about the intervening sequence and are particularly useful for de novo transcriptome construction and detecting structural variants, such as indels.
Stranded reads
RNA-seq data can come in either stranded or unstranded varieties. If the data is unstranded, the strand from which the fragment was transcribed cannot be identified directly from the sequence. Furthermore, because the RNA-seq protocol usually involves forming double stranded cDNA for ease of sequencing, the returned sequence is just as likely to be that of the reverse complement of the source DNA sequence as the original DNA sequence. In practical terms, this means that while half the reads map to the forward strand and half the reverse, this mapping does not contain any information about which strand the RNA was transcribed from.
Stranded RNA-seq data on the other hand preserves strand information, making it possible to identify which strand the RNA was transcribed from.
Protocols
Typical workflow
Spliced Mapping
TopHat
Tophat is a tool that acts as a fast splice junction mapper for RNA-Seq reads. Tophat maps RNA-Seq reads to a mammalian genome using Bowtie, an ultrahigh-throughput short read aligner that utilizes a Burrows-Wheeler index. After the reads are aligned by Bowtie, Tophat analyzes the mapping results to identify splice junctions between exons. Tophat is one of the few tools effective for dealing with intronic regions, which are more prevalent in reads that are greater than 50 bp in length.we can use HISAT instead of tophat
GSNAP
MapSplice
File formats
GFF/GTF
SAM/BAM
Creating a dataset
Cufflinks
MISO
GSTRUCT
Reference datasets
Human
The Genome Reference Consortium provides a human reference genome that is constructed from several individuals from a diverse population. And the quality of the reference is continually improved with the correction of extant assembly errors, including but not limited to incorrect mixing the haplotype structure among the individuals used.
CCDS
The Consensus CDS (CCDS) project is a collaborative effort to identify a core set of human and mouse protein coding regions that are consistently annotated and of high quality. The annotation are consensus among NCBI's RefSeq, EBI's Ensembl and Sanger's Havana.
GENCODE
GENCODE is a sub-project of the ENCODE scale-up project to annotate all evidence-based gene features in the entire human genome.
HAVANA
The HAVANA group at the Sanger Institute manually annotate the human genome, providing comprehensive annotation for full complexity of gene loci and features that may not be well catered by an automated annotation system.[3]
RefSeq
RefSeq is a reference sequence annotation service provided by the NCBI.[4]
UCSC genes
UCSC Known genes are constructed by a fully automated process, based on protein data from Swiss-Prot/TrEMBL (UniProt) and the associated mRNA data from Genbank.[5]
Viewing datasets
Browsers
Comparing datasets
SEQwiki content dump
What does the data look like?
What can be done with RNASeq data
In order to analyze your RNASeq data, there are different aims which require different preprocessing steps.
Calling DE genes
If you want to call differentially expressed genes it is mandatory for many BioConductor packages that you do not convert your read count to e.g. rpkm but use the raw read count. The reason is that the statistical model uses the original read count data. (Less reads mean more short noise, whereas a lower rpkm value could still be associated to many reads e.g. in the case of a very long gene). Please also bear in mind that even if you use RNASeq data you still need biological replicates.
Also you will want to use software packages that are made for RNAseq (i.e count data) such as DESeq, bayseq, NBPSeq or edgeR, which all use a negative binomial model.
A typical workflow might look like this
- quality control your data
- Mapping Reads (see the howto)
- Counting Reads per feature
- statistical analysis
Counting/summarizing reads
After having aligned reads to your genome you need to summarize the reads. This might be a crucial step, as there are many different ways to do summarization. One way could be to be very stringent and only count completely unambiguous reads, however in downstream analyses this needs to be taken into account. One could summarize the data by using e.g. HTSeq count[6] or the Bioconductor Iranges module
Statistical Analysis
When testing for differential expression, one should likely take a package modelling count data. In Bioconductor, options include DESeq,[7] edgeR and bayseq.
Differential exon usage
Besides overall changes in expression of a gene, there can be differentially abundant of isoforms or specific exons. Testing for differentially abundant exons can be done using DEXSeq,[8] see also the Genome Research paper.[9]
Category enrichment (e.g. GO enrichment)
As with all enrichment analyses, the categories that you find might be confounded by detection power. With RNASeq, the power for detecting differential expression (at the same type-I error, e.g. the same adjusted p-value) is higher for genes with more counts. For example, if all photosynthesis genes were highly expressed, you would have a higher chance to find any of these to be differentially expressed than other genes. Now, if you found photosynthesis genes enriched in your list of differentially expressed genes, that might be just because they were easier to find.
The authors of the Goseq package in Bioconductor noted that longer genes tend to generate more counts, and they provide software for doing GO category enrichment analysis that is able to adjust either for gene length, or directly for expression strength.
Estimating expression height
A typical workflow might look like this
- Mapping Reads
- Counting Reads per feature
- normalizing for gene length
Finding novel genes or splice variants
Quality Control (QC)
There are many possible avenues for performing checks for quality on sequencing data. Some popular options are: FastQC,[10] DNAA[11] or you can do it in R or use HTSeq
Although more complex quality metrics can be used, a basic check that the sequence composition doesn't vary too greatly along the length of the reads and the quality scores do not dip too low are a good place to start. These checks (as well as several others) can be performed by loading the fastq files into the fastQC program.
Mapping the reads
Unless your aim is to do de novo transcriptome assembly, the first step of any analysis will be to align your millions of short reads to a reference of some kind. This is usually the reference genome for the species from which the RNA was extracted. There are many different aligners for performing short-read alignment to a reference (lists are available at http://en.wikipedia.org/wiki/List_of_sequence_alignment_software and http://seqanswers.com/wiki/Special:BrowseData/Bioinformatics%20application?Bioinformatics_method=Mapping). Each has specific advantages and disadvantages, usually involving a trade-off between speed and sensitivity. For the purposes of this guide we will use the aligner BOWTIE because it is actively developed, fast, widely used and supports both base space and colour space reads.
As a starting point, I will assume you have the following files:
For single end reads:
sample.fa or sample.cfa (cfa for colour space, fa for base space)
For paired end reads:
sample_pair1.fa and sample_pair2.fa or sample_pair1.cfa and sample_pair2.cfa (cfa for colour space, fa for base space)
Obtaining the reference
In order to map the short reads to a reference genome, that genome has to be turned into an index which can be used by BOWTIE. A number of prebuilt indexes for common genomes can be downloaded from the BOWTIE website. If your genome is not available you will have to construct the index yourself from a fasta file containing your reference genome, these can be obtained from UCSC via FTP . This is accomplished using the command:
bowtie-build reference.fa reference_name
The argument "reference_name" is a unique identifier that will be used to refer to this reference genome from now on. If your reads are colour space, construct a colour space index by adding the -C command:
bowtie-build -C reference.fa reference_name
This command will output 6 files named reference_name.1.ebwt, reference_name.2.ebwt, reference_name.3.ebwt, reference_name.4.ebwt, reference_name.rev.1.ebwt, and reference_name.rev.2.ebwt.
Irrespective of how you obtain the index, in order for BOWTIE to use it, the six files mentioned above need to be placed in the BOWTIE indexes directory. If you're not sure what your indexes directory is, it is pointed to by the environmental variable BOWTIE_INDEXES, so:
echo $BOWTIE_INDEXES
will display the path where you should put the index files.
If you wish to use an aligner other than BOWTIE, you will also have to build an index from the reference genome. Refer to the documentation for your preferred aligner for more information.
Aligning reads to the reference
Having constructed the reference into an index that BOWTIE can use, we now want to align our data to this index. BOWTIE offers a wealth of command line options that can be used to adjust the alignment algorithm and how it handles input/output. These command line options are described in detail in the BOWTIE manual, but there are a few flags that are commonly used in the analysis of RNA-seq and bear mentioning here.
The --sam or -S tells BOWTIE to output the results of the alignment in SAM format instead of the BOWTIE format. SAM[12] is rapidly becoming the standard for reporting short read alignment and is supported by a wide range of downstream analysis tools. Unless you have a very good reason not to, this flag should always be specified.
The --best flag tells BOWTIE to guarantee that the alignment that it reports has the fewest number of mismatches to the reference out of all matches found. It also performs a few other desirable functions such as removing strand bias.[13] The trade-off for these benefits is that --best is slightly slower, but this difference is negligible in almost all instances. Note that --best does not apply to paired-end reads. This flag should be enabled unless speed is a major consideration.
Each base has a quality score associated with it which is reported on the PHRED scale (http://en.wikipedia.org/wiki/Phred_quality_score) where lower scores mean less confidence in the accuracy of the base call. For each candidate alignment, BOWTIE adds up the quality scores at bases which don't match the reference. Any match location that has a sum of mismatch quality scores greater than -e is deemed invalid and not reported. The default value of -e of 70 was optimized when read lengths tended to be shorter (~30bp), but is not appropriate for longer read lengths commonly used today. Furthermore, any true biological variation from the reference (such as SNPs) will in theory have a high quality score. Therefore, reads with SNPs will score very poorly on the sum of mismatching quality scores metric. For all these reasons, it is advised that the user increase -e beyond the default unless the reference is known to be an excellent representation of the biological source of RNA and the number of errors in the read is small.
The -p flag sets the number of simultaneous threads to use. As short read alignment is effectively infinitely parallelizable, set this to the number of CPU cores available.
With these considerations in mind, we map our short read data to our reference using the command
bowtie -p 8 --sam --best reference_name sample.fa aligned_reads.sam
If using colour space, the -C flag needs to be added.
bowtie -p 8 --sam -C --best reference_name sample.cfa aligned_reads.sam
If your data is paired end the two paired files need to be specified using the -1 and -2 flags
bowtie -p 8 --sam reference_name -1 sample_pair1.fa -2 sample_pair2.fa aligned_reads.sam
More complex alignment
The fragments of cDNA that are being sequenced originate not from the genome, but from the transcriptome. The transcriptome is formed by combining exons from the genome, which is why mapping to the genome is a good approximation. However, in doing so the ability to map any read that crosses exon-exon boundaries is lost. The longer the reads, the more this becomes an issue as a read is more likely to cross a boundary and be rendered unmappable. Depending on the desired downstream analysis, this lost coverage at exon boundaries may or may not be a problem.
Exon Junction libraries
It is possible to build exon junction libraries from known annotated exons and then try and map those reads that fail to map against this sequence. Such an approach is only capable of capturing reads that span known annotations, limiting its utility and biasing the results towards well annotated genes/genomes. In order to do this, a new reference needs to be constructed that contains both the reference genome, as well as the new exon-junction sequence. It is important that the original genomic reference still be included in the reference, even if you are only mapping those reads that failed to align to the genome, so as the reads can compete amongst all possible mapping locations when determining alignment. You also need to decide how much sequence to take from either side of the exon-exon boundary. The amount of sequence from each exon must be less than the read length, otherwise any read which falls near an exon boundary, but not over it, will map to two locations, the exon junction library and the genome itself. This means that if you intend to trim your reads, the amount of sequence you take from each side of the exon junction must me less than the length of the trimmed read. Finally, you need to decide what combinations of exon-exon joinings you are going to consider. Do you only consider splicing within genes, within chromosomes, without exon reorientation? All of these things occur with different frequencies in different samples and the more possibilities you consider the greater the computational complexity.
To construct the junction library you first need a genome annotation. These can be downloaded from the UCSC table browser. You can then either write your own tools to create a fasta file with all the exon junctions, or use an existing tool, such as the "Make Splice Junction Fasta" application included in the USeq software package.[14] Once you have a fasta file containing your exon-junction libarary you need to combine it with the fasta file for your reference genome.
cat reference.fa junctions.fa >reference_and_junctions.fa
Then build a new bowtie index as described above
bowtie-build reference_and_junctions.fa junction_lib
Finally, you map the reads in the same way as to the reference genome. For example,
bowtie -p 8 --sam --best junction_lib sample.fa aligned_reads.sam
Further options
If this approach still does not map a reasonable number of reads, there are other alternative approaches that can be explored, at even greater computation cost. For example, you may try to estimate splice junctions from the data itself, using "De-novo" splice junction finders. There are many tools that attempt to do this, some examples include TopHat,[15] SplitSeek,[16] PerM,[17] and SOAPsplice (formerly SOAPals).[18] Full de novo assembly of the transcriptome is also a possibility, although very high coverage is required for this to work well. Paired end data is of huge benefit to this task as either end of a fragment mapping to a different exon is very strong evidence for a splice junction. A list of such tools can be found in Table 1 of the review article.[1]
A recent paper in genome biology further suggested a automatic framework to easily benchmark and choose a mapper given key characteristics of the data sets (e.g. reference genome, SNP rate, read length, etc.). Teaser is available as an online tool[19] or you can download and customize your own version.[20]
Differential Expression
A common use of expression assays is to look for differences in expression levels of genes or other objects of interest between two experimental conditions, such as a wildtype vs knockout. In order to do this we need to transform the data from a list of reads mapping to genomic coordinates into a table of counts. The strategy we employ here is to load the short reads into R using the Rsamtools package and then count the number of reads overlapping some annotation object, which is usually something like a collection of genes downloaded from the UCSC. Once transformed, a test can be performed to look for statistically significant differences in expression level.
Summarization of reads
Compressing aligned reads using SAMtools
In order to be able to load millions of aligned reads into memory in R, we need to create a binary compressed version of our human readable SAM output. This file will contain all the same information, but have a much smaller memory footprint as well as being quickly searchable. To create such files, we first need to install SAMtools.[12] Next we need to construct an index of the reference, using the fasta file. This is done by executing:
samtools faidx reference.fa
Which creates a file reference.fa.fai. Next we convert SAM to BAM. BAM files contain all the same information as SAM files, but are compressed to be more space efficient and searchable.
samtools import reference.fa.fai aligned_reads.sam aligned_reads.bam
Finally we need to create an index of the reads so they can be quickly searched. In order to do this we first need to sort the BAM file.
samtools sort aligned_reads.bam aligned_reads_sorted
This will create aligned_reads_sorted.bam, which we now index.
samtools index aligned_reads_sorted.bam
Which creates the index file aligned_reads_sorted.bam.bai.
If you don't have the original fasta file for the reference because you downloaded a prebuilt index from the BOWTIE website (or because you lost the fasta file after making your own), you can rebuild the source fasta file by running the following.
bowtie-inspect reference_name>reference.fa
Working with BAM files in R
Our goal is to use R to summarize the reads by genes for each sample. To this end, we will use our newly created compressed representation of the short reads (the sorted, indexed, BAM file).
Fetching gene information
The first thing we need to do is to define the location of genes in chromosome coordinates. To do this, we use the GenomicFeatures package.[21] This package allows us to download gene information from the UCSC genome browser using the following commands:
library(GenomicFeatures) txdb=makeTranscriptDbFromUCSC(genome='hg19',tablename='ensGene')
Various genomes and gene IDs are available, but as an example we will use the latest human genome and ENSEMBL gene IDs. The variable "txdb" now contains all the information we need, but in order to do anything with it we need to do some processing.
tx_by_gene=transcriptsBy(txdb,'gene')
This produces a GRangesList object which is a list of GRanges objects, where each GRanges object is a gene and the entries are the genomic coordinates of its transcripts. We are going to work out which reads overlap which genes using the countOverlaps function to overlap this object with the object containing the short reads.
Although we are choosing to summarize by including all reads that fall within a gene here, the procedure will work the same for any GRanges or GRangesList object. For example, if you wished to only include reads that overlap exons, you could create a different GRangesList object.
ex_by_gene=exonsBy(txdb,'gene')
Summarizing reads in R
First we load the aligned_reads_sorted.bam file into R.
library(Rsamtools)
reads=readBamGappedAlignments("aligned_reads_sorted.bam")
Checking compatibility of annotations and reads
Before we try and compare the reads to the annotation, we first need to do a few checks to make sure that everything will work OK. If your RNA-seq data did not come with strand information, than we cannot know which strand the read was transcribed from. However, the mapping process will map it to one strand or the other (in theory, both are equally likely), thus the reads will have a strand artificially allocated to them. When we count the number of reads overlapping a gene (or other feature), only those reads that map to the strand the gene is on will count and roughly half our reads will be lost. To avoid this we need to set the reads strand value to "*" (unknown).
Furthermore, it will often be the case that the chromosome names used by the alignment software (which are ultimately determined by the chromosome names in the fasta file for the reference genome) will differ from those given in the annotation. In order for the comparison function to work, these names need to be converted to the same naming convention. It is easy to check if the names match by listing all the chromosomes for both reads and annotations. Remember, our annotation data is stored in "tx_by_gene" and our short reads are stored in "reads".
#The annotations have chromosomes called names(seqlengths(tx_by_gene)) #The reads have chromosomes called as.character(unique(rname(reads)))
If the chromosome names are the same (or are the same for the ones you care about), then no name conversion is needed. If on the other hand they differ, we need to change either the reads or the annotations naming convention. It turns out that it is usually easier to change the names of the reads, but the procedure is the same regardless. This is best illustrated by an example. Suppose you have this situation:
#The annotations have chromosomes called > names(seqlengths(tx_by_gene)) [1] "chr1" "chr10" "chr11" "chr12" "chr13" [6] "chr13_random" "chr14" "chr15" "chr16" "chr17" [11] "chr17_random" "chr18" "chr19" "chr1_random" "chr2" [16] "chr3" "chr3_random" "chr4" "chr4_random" "chr5" [21] "chr5_random" "chr6" "chr7" "chr7_random" "chr8" [26] "chr8_random" "chr9" "chr9_random" "chrM" "chrUn_random" [31] "chrX" "chrX_random" "chrY" "chrY_random"
#The reads have chromosomes called >as.character(unique(rname(reads))) [1] "10.1-129993255" "11.1-121843856" "1.1-197195432" "12.1-121257530" [5] "13.1-120284312" "14.1-125194864" "15.1-103494974" "16.1-98319150" [9] "17.1-95272651" "18.1-90772031" "19.1-61342430" "2.1-181748087" [13] "3.1-159599783" "4.1-155630120" "5.1-152537259" "6.1-149517037" [17] "7.1-152524553" "8.1-131738871" "9.1-124076172" "MT.1-16299" [21] "X.1-166650296" "Y.1-15902555"
So we need to convert the read chromosome names "NO.1-Length" to the annotation name "chrNO".
new_read_chr_names=gsub("(.*)[T]*\\..*","chr\\1",rname(reads))
If you are not familiar with regular expressions, refer to the help file for gsub in R. new_read_chr_names will now contain the read chromosome names, converted to the same format as the annotation object tx_by_genes.
Now we can fix both the chromosome name and strand problem simultaneously, by building a GenomicRanges object from each of the read objects. If you have unstranded RNA-seq data and need to convert chromosomes, we run:
reads=GRanges(seqnames=new_read_chr_names,ranges=IRanges(start=start(reads),end=end(reads)),
strand=rep("*",length(reads)))
If we just want to convert chromosome names.
reads=GRanges(seqnames=new_read_chr_names,ranges=IRanges(start=start(reads),end=end(reads)),
strand=strand(reads))
If we just want to make each read be ambiguous with respect to strand.
reads=GRanges(seqnames=rname(reads),ranges=IRanges(start=start(reads),end=end(reads)),
strand=rep("*",length(reads)))
Note that if you mapped reads to an exon-junction library, every exon junction will have its own "chromosome". If you wish these junctions reads to be included in the summarization, you will have to convert each of them to genomic coordinates. As each read comes from two distinct genomic locations (either side of the exon junction), you will have to make a decision about how you are going to assign each read a genomic coordinate.
Counting the number of reads
Finally, we get the number of reads that overlap with each gene (or whatever else you're interested in).
counts=countOverlaps(tx_by_gene,reads)
"counts" will now contain a numeric vector, where the ith entry is the number of reads that overlap the ith gene in "tx_by_gene".
Differential Expression Testing
As our aim is to compare conditions, we will have more than one lane of reads, possibly several for each condition. Using the procedure outlined in the previous section, we can count the number of reads that overlap a feature of interest, such as genes, in each experimental condition, for each replicate. Next we combine them into a table of counts.
toc=data.frame(condition1_rep1=counts1.1,condition1_rep2=counts1.2,
condition2_rep1=counts2.1,condition2_rep2=counts2.2,stringsAsFactors=FALSE)
rownames(toc)=names(tx_by_gene)
etc. for as many conditions and replicates as are available. Here the convention is countsn.m is the vector containing the number of reads from replicate m of condition n that overlap the genes given by tx_by_gene.
Normalization
It has been shown that a small number of highly expressed genes can consume a significant amount of the total sequence. As this can change between lanes and experimental condition, along with library size, it is necessary to perform some kind of between sample normalization when testing for differential expression. The choice of normalization is not independent of the test used to determine if any genes are significantly differentially expressed (DE) between conditions. For example, quantile normalization produces non-integer counts, making tests based on the assumption of count data such as the widely used Poisson or Negative Binomial models inapplicable. We choose to use the scaling factor normalization method as it preserves the count nature of the data and has been shown to be an effective means of improving DE detection.[22]
To perform the normalization and the test for differential expression, we will use the R package edgeR,[23] although there are other options available. We can now calculate the normalization factors using the TMM method.[22]
library(edgeR) norm_factors=calcNormFactors(as.matrix(toc))
Statistical testing
Next, we have to create a DGE object used by edgeR. The scaling factor calculated using the TMM method is incorporated into the statistical test by weighting the library sizes by the normalization factors (which are then used as an offset in the statistical model).
DGE=DGEList(toc,lib.size=norm_factors*colSums(toc),group=rep(c("Condition1","Condition2"),c(2,2)))
The group variable identifies which columns in the table of contents come from which experimental condition or "group". To perform the statistical test for significance, we first estimate the common dispersion parameter
disp=estimateCommonDisp(DGE)
Finally, we calculate the p-values for genes being DE
tested=exactTest(disp)
Gene Set testing (GO)
To accurately test sets of genes for over representation amongst DE genes using RNA-seq data, we need to use a method which takes into account the biases particular to this technology. The GOseq package[24] is one such method for accounting for certain RNA-seq specific biases when performing GO (and other gene set based tests) analysis.[25]
First we must format the output of edgeR to be read by goseq. We call any gene with a Benjamini-Hochberg FDR of less than .05 DE.
library(goseq) genes = as.integer(p.adjust(tested$table$p.value, method = "BH") < 0.05) names(genes) = row.names(tested$table)
Next, we calculate a probability weighting function, correcting for length bias, a technical bias present in all forms of RNA-seq data.[25]
pwf=nullp(genes,'hg19','ensGene')
If we'd instead wanted to correct for total read count bias, we would calculate the pwf using the number of counts from each gene as follows:
pwf=nullp(genes,bias.data=rowsum(counts[match(names(genes),rownames(counts))]))
Finally, we calculate the p-value for each GO category being over represented amongst DE genes.
GO.pvals=goseq(pwf,'hg19','ensGene')
Example 1: Differential Expression and GO Term Analysis: Li Prostate cancer data set
This section provides an easy to follow example to illustrate the analysis pipeline outlined above.
Description
This data set compares prostate cancer LNcap cell lines with and without treatment by the testosterone like hormone androgen.[26] The sequencing was done using the Illumina GA I and produced 36 bp, single end, unstranded reads. The output from the machine are 7 files (each from a different sequencing lane):
untreated1.fa
untreated2.fa
untreated3.fa
untreated4.fa
treated1.fa
treated2.fa
treated3.fa
Note that this data set is slightly unusual in that the quality scores are missing from the reads. Therefore, we will have to keep this in mind when doing the analysis.
Quality Control
It's published data, so that's a pretty good quality control one would hope...
Sequence alignment
Building the reference
The first step in the pipeline is to align all the reads to a reference. As this data is taken from human LNcap cells, the latest build of the human genome is an obvious choice. We have installed BOWTIE (version 0.10.0) using all the standard options. There is a prebuilt copy of the human index available from the BOWTIE website, however, to illustrate building a genome from scratch we instead download the .fa files for the genome from the UCSC. We create a working directory containing the 7 RNA-seq data files and the file chromFA.tar.gz downloaded from University of California - Santa Cruz. The file chromFA.tar.gz contains the sequence of all human chromosomes, including the unallocated contigs. To make a BOWTIE index we need to concatenate them into a single file. We will exclude all the contigs from our fasta file.
tar -zxvf chromFA.tar.gz
We only want chr1-22.fa, chrX.fa, chrY.fa and chrM.fa, so delete everything else:
rm chr*_*.fa
Now we concatenate the desired files (it is useful to have the reference both in one chromosome per file and one file per genome formats, although the BOWTIE index can be made from either).
cat chr*.fa>hg19.fa
We build the BOWTIE index and name it hg19.
bowtie-build hg19.fa hg19
And move the BOWTIE index files to the appropriate location for BOWTIE to find them.
mv *.ebwt $BOWTIE_INDEXES
Aligning the reads
Having constructed the BOWTIE index for the human genome, we now proceed to map the reads from each lane. We want to use the --best and --sam flags. At this point we recall that our data lacks quality information for the reads. Therefore, we use the -v 3 option which ignores quality scores when aligning reads to the genome.
for src_fastafile in *treated*.fa
do
bowtie -v 3 -p 8 --best --sam hg19 ${src_fastafile} ${src_fastafile%%.fa}.sam
done
The for loop is just for convenience and is equivalent to writing:
bowtie -v 3 -p 8 --best --sam hg19 untreated1.fa untreated1.sam bowtie -v 3 -p 8 --best --sam hg19 untreated2.fa untreated2.sam bowtie -v 3 -p 8 --best --sam hg19 untreated3.fa untreated3.sam bowtie -v 3 -p 8 --best --sam hg19 untreated4.fa untreated4.sam bowtie -v 3 -p 8 --best --sam hg19 treated1.fa treated1.sam bowtie -v 3 -p 8 --best --sam hg19 treated2.fa treated2.sam bowtie -v 3 -p 8 --best --sam hg19 treated3.fa treated3.sam
BOWTIE will output 7 SAM files containing the aligned reads. To count the fraction of aligned reads we run the following command at the shell (this information is also reported directly by BOWTIE, but it is useful to be able to calculate it yourself if need be).
awk '$3!="*"' untreated1.fa|wc -l wc -l untreated1.fa
The first command prints the number of reads that have mapped in the BOWTIE output file, the second outputs 4 times the number of reads in the input file (because the fasta format is for 4 lines per read).
We will ignore those reads that cross exon-exon boundaries and continue with the analysis.
Summarization of reads
Converting to BAM
In order to summarize our aligned reads into genes, we first have to convert the SAM output of BOWTIE into the compressed, index BAM format. First we need to create an index for the human genome in the samtools format. For this we need the fasta file for the reference (which should be the same that was used to create the index for aligning the reads), which we already have, but we will reconstruct it from the BOWTIE index anyway.
bowtie-inspect hg19>hg19.fa
Now we construct the samtools index. The following command produces a .fai file which we can use to convert the SAM files to BAM files.
samtools faidx hg19.fa
Next, we convert all 7 SAM files to BAM files.
for SAM in *.sam
do
#Convert to BAM format
samtools import hg19.fa.fai ${SAM} ${SAM%%.sam}.bam
#Sort everything
samtools sort ${SAM%%.sam}.bam ${SAM%%.sam}_sorted
#Create an index for fast searching
samtools index ${SAM%%.sam}_sorted.bam
#Delete temporary files
rm ${SAM%%.sam}.bam
done
Again, the bash for loop is just for convenience, the above is equivalent to running the following for all 7 files:
samtools import hg19.fa.fai untreated1.sam untreated1.bam samtools sort untreated1.bam untreated1_sorted samtools index untreated1_sorted.bam rm untreated1.bam
Processing in R
Next we need to load the sorted, indexed, BAM files into R. As we have unstranded RNA-seq, we need to make the strand designator for each read ambiguous (which is done by setting it to "*"). After starting R we run,
library(Rsamtools)
#Create a list of bam file object containing the short reads
bamlist=list()
src_files=list.files(pattern="*_sorted.bam$")
for(filename in src_files){
#Since we do not know which strand the reads were originally transcribed,
#so set the strand to be ambiguous
tmp=readBamGappedAlignments(filename)
bamlist[[length(bamlist)+1]]=GRanges(seqnames=rname(tmp),
ranges=IRanges(start=start(tmp),end=end(tmp)),
strand=rep("*",length(tmp)))
}
names(bamlist)=src_files
Having loaded the files into R, we next need to create an annotation object. Since we are using hg19, we can readily download one from the UCSC using the GenomicFeatures package. We choose to use the ENSEMBL gene annotation.
library(GenomicFeatures) txdb=makeTranscriptDbFromUCSC(genome="hg19",tablename="ensGene")
We want to compare genes for differential expression, so we will summarize by gene and we choose to count all reads that fall within the body of the gene (including introns) as counting towards a genes count.
tx_by_gene=transcriptsBy(txdb,"gene")
Finally, we count the number of reads that fall in each gene for each lane and record the results in a table of counts.
#Initialize table of counts
toc=data.frame(rep(NA,length(tx_by_gene)))
for(i in 1:length(bamlist)){
toc[,i]=countOverlaps(tx_by_gene,bamlist[[i]])
}
#Fix up labels
rownames(toc)=names(tx_by_gene)
colnames(toc)=names(bamlist)
Differential Expression testing
Having finally obtained a table of counts, we now want to compare the treated and untreated groups and look for any statistically significant differences in the number of counts for each gene. We will do this using the negative binomial model used by edgeR.
Normalization
We calculate appropriate scaling factors for normalization using the TMM method with the first lane as the reference.
library(edgeR) norm_factors=calcNormFactors(as.matrix(toc))
The counts themselves are not changed, instead these scale factors are used as an offset in the negative binomial model. This is incorporated in the DGE list object required by edgeR.
DGE=DGEList(toc,lib.size=norm_factors*colSums(toc),group=gsub("[0-9].*","",colnames(toc)))
Statistical Test
Next we calculate a common dispersion parameter which represents the additional extra Poisson variability in the data.
disp=estimateCommonDisp(DGE)
Which allows us to calculate p-values for genes being differentially expressed.
tested=exactTest(disp)
Gene Ontology testing
In order to test for over represented GO categories amongst DE genes, we first have to pick a cutoff for calling genes as differentially expressed after applying multiple hypothesis correction. We choose the ever popular cutoff for significance of .05
library(goseq) #Apply benjamini hochberg correction for multiple testing #choose genes with a p-value less than .05 genes=as.integer(p.adjust(tested$table$p.value,method="BH") <.05) names(genes)=row.names(tested$table)
Now we calculate the probability weighting function, which quantifies the length bias effect.
pwf=nullp(genes,"hg19","ensGene")
Finally, we calculate the p-values for each GO category being over represented amongst DE genes.
GO.pvals=goseq(pwf,"hg19","ensGene")
Example 2 Differential Expression: Di Arabidopsis pathogen data
Donwnload the six fastq.gz files from here: http://www.ebi.ac.uk/ena/data/view/SRP004047&display=html
These files are from an Arabidopsis study using three replicates each from infected and mock infected plants. This is the data set underlying the NBPSeq R package.
Quality Control
Download FastQC and open the files in FastQC one by one. You can open the files by using File->Open.
As of Version 0.94. if you are on Windows use the Linux version and double-click run_fastqc.bat
You will see a very wiggly line for the first library. If you just look at the peaks and note the sequence you will see the pattern AAGAGCTCGTATGC starting at the green plateau towards the right. This is an illumina adapter sequence, which you will also see in the overrepresented counts tab.
Mapping Reads
Download the Arabidopsis genome release from here
ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/other_datasets/CURRENT/arabidopsis.seq.gz It doesn't include mitochondrial or chloroplast sequences but is good enough for the tutorial purpose
unzip all read files
Using bowtie an index needs to be built
Unix systems bowtie-build arabidopsis.seq arabidopsis
Windows C:\Prog\bowtie-0.12.7\bowtie-build.exe arabidopsis.seq arabidopsis
Alternatively, you can download the provided index file from the bowtie website if you work with one of the supported organisms.
Now you can map reads using bowtie. Bowtie has many options and you better check them. Here we tell it to use two processors (-p 2) to report SAM based alignements -S for all (-a) alignment having maximally one mismatch (-v 1). We further restrict this based on the fact that only each read providing more than one valid alignment should be discarded (-m 1)
bowtie -p 2 -S -a -m 1 -v 1 arabidopsis SRR074262.fastq aligned_074262 bowtie -p 2 -S -a -m 1 -v 1 arabidopsis SRR074263.fastq aligned_074263 bowtie -p 2 -S -a -m 1 -v 1 arabidopsis SRR074264.fastq aligned_074264 bowtie -p 2 -S -a -m 1 -v 1 arabidopsis SRR074284.fastq aligned_074284 bowtie -p 2 -S -a -m 1 -v 1 arabidopsis SRR074285.fastq aligned_074285 bowtie -p 2 -S -a -m 1 -v 1 arabidopsis SRR074286.fastq aligned_074286
C:\Prog\bowtie-0.12.7\bowtie.exe -p 2 -S -a -m 1 -v 1 arabidopsis SRR074262.fastq aligned_074262
This needs to be repeated for all 6 libraries.
Output SRR074262.fastq
- reads processed: 9619406
- reads with at least one reported alignment: 5034111 (52.33%)
- reads that failed to align: 4132070 (42.96%)
- reads with alignments suppressed due to -m: 453225 (4.71%)
Reported 5034111 alignments to 1 output stream(s)
Output SRR074263.fastq
- reads processed: 4199495
- reads with at least one reported alignment: 2298490 (54.73%)
- reads that failed to align: 1701822 (40.52%)
- reads with alignments suppressed due to -m: 199183 (4.74%)
Reported 2298490 alignments to 1 output stream(s)
Output SRR074264.fastq
- reads processed: 4810892
- reads with at least one reported alignment: 2730242 (56.75%)
- reads that failed to align: 1852040 (38.50%)
- reads with alignments suppressed due to -m: 228610 (4.75%)
Reported 2730242 alignments to 1 output stream(s)
Output SRR074284.fastq
- reads processed: 4763394
- reads with at least one reported alignment: 2622118 (55.05%)
- reads that failed to align: 1908441 (40.06%)
- reads with alignments suppressed due to -m: 232835 (4.89%)
Reported 2622118 alignments to 1 output stream(s)
Output SRR074285.fastq
- reads processed: 8217239
- reads with at least one reported alignment: 4515790 (54.96%)
- reads that failed to align: 3206578 (39.02%)
- reads with alignments suppressed due to -m: 494871 (6.02%)
Reported 4515790 alignments to 1 output stream(s)
Output SRR074286.fastq
- reads processed: 3776979
- reads with at least one reported alignment: 2014498 (53.34%)
- reads that failed to align: 1506142 (39.88%)
- reads with alignments suppressed due to -m: 256339 (6.79%)
Reported 2014498 alignments to 1 output stream(s)
Summarizing Reads
If you want to use HTSeq-count[6] and are on Fedora or CentOS you will have to go through some extra effort, as HTSeq uses python 2.6 and Fedora and CentOS only come with python 2.4 installed. (Before you do any of the following ask your sysadmin if this is ok)
here are some suggestions http://stackoverflow.com/questions/1465036/install-python-2-6-in-centos My favorite one is to use EPEL (see below), buy YMMV.
rpm -Uvh http://download.fedora.redhat.com/pub/epel/5/i386/epel-release-5-4.noarch.rpm yum install python26 yum install python26-numpy yum install python26-numpy-devel
Then continue as detailed on the website but type
python26 setup.py build python26 setup.py install
instead of
python setup.py build python setup.py install
---
Now we are ready to download the gff file: ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff
This file does not contain RNA genes and transposable elements.
Now the parent ID probably works for some use cases of HTSeq. If you needed a true GTF file you could use the perl script mentioned here at the bottom. If we named it celeste_script.pl here would be what to execute. (The reason we can't use -i Parent is that otherwise an exon might belong to two splice variants e.g. AT1G01040.1 and AT1G01040.2 and not be counted as we want to use intersection-strict)
cat TAIR10_GFF3_genes.gff |perl celeste_script.pl > TAIR10_GTF.gtf
and now finally
htseq-count -m intersection-strict -s no aligned_074262 TAIR10_GTF.gff > counts_074262 htseq-count -m intersection-strict -s no aligned_074263 TAIR10_GTF.gtf > counts_074263 htseq-count -m intersection-strict -s no aligned_074264 TAIR10_GTF.gtf > counts_074264 htseq-count -m intersection-strict -s no aligned_074284 TAIR10_GTF.gtf > counts_074284 htseq-count -m intersection-strict -s no aligned_074285 TAIR10_GTF.gtf > counts_074285 htseq-count -m intersection-strict -s no aligned_074286 TAIR10_GTF.gtf > counts_074286
Now we start getting something we can use
head counts_074262
AT1G01010 75 AT1G01020 73 AT1G01030 33 AT1G01040 109 AT1G01046 0 AT1G01050 41 AT1G01060 10 AT1G01070 2 AT1G01073 0 AT1G01080 281
head counts_074263
AT1G01010 35 AT1G01020 34 AT1G01030 18 AT1G01040 52 AT1G01046 0 AT1G01050 51 AT1G01060 5 AT1G01070 19 AT1G01073 0 AT1G01080 212
head counts_074264
AT1G01010 80 AT1G01020 46 AT1G01030 45 AT1G01040 50 AT1G01046 0 AT1G01050 69 AT1G01060 13 AT1G01070 35 AT1G01073 0 AT1G01080 226
head counts_074284
AT1G01010 45 AT1G01020 50 AT1G01030 33 AT1G01040 65 AT1G01046 0 AT1G01050 50 AT1G01060 0 AT1G01070 7 AT1G01073 0 AT1G01080 178
head counts_074285
AT1G01010 35 AT1G01020 43 AT1G01030 26 AT1G01040 72 AT1G01046 0 AT1G01050 51 AT1G01060 3 AT1G01070 6 AT1G01073 0 AT1G01080 531
head counts_074286
AT1G01010 61 AT1G01020 27 AT1G01030 36 AT1G01040 25 AT1G01046 0 AT1G01050 25 AT1G01060 16 AT1G01070 20 AT1G01073 0 AT1G01080 128
Analysis of differentially expressed genes
start R
using DESeq (now refer to the vignette)
R
In R:
library(DESeq)
setwd("where the data is")
c1<-read.table("counts_074284",row.names=1)
c2<-read.table("counts_074286",row.names=1)
c3<-read.table("counts_074262",row.names=1)
c4<-read.table("counts_074263",row.names=1)
c5<-read.table("counts_074264",row.names=1)
c6<-read.table("counts_074285",row.names=1)
counts<-cbind(c1,c2,c3,c4,c5,c6)
counts<-counts[-c(32679:32683),] #remove the more general lines
colnames(counts)<-c("P1","P2","P3","M1","M2","M3")
design <- rep (c("P","Mo"),each=3)
de <- newCountDataSet(counts, design)
de <- estimateSizeFactors( de)
# de <- estimateVarianceFunctions( de ) # This function has been removed. Use 'estimateDispersions' instead.
de <- estimateDispersions( de )
res <- nbinomTest( de, "P", "Mo")
How many genes are significant?
sum(na.omit(res$padj<0.05))
References
- ↑ a b Oshlack, A.; Robinson, M.D.; Young, M.D. (2010). "From RNA-seq reads to differential expression results". Genome Biology. 11 (12): 220. doi:10.1186/gb-2010-11-12-220. PMC 3046478. PMID 21176179.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ "Applied Biosystems SOLiD 3 System: SOLiD Analysis Tools (SAT) V3.0 User Guide" (PDF). Life Technologies Corporation. April 2009. p. 212. Retrieved 30 April 2016.
- ↑ "Vertebrate Annotation - Sanger Institute". Sanger Institute. Retrieved 30 April 2016.
- ↑ "RefSeq: NCBI Reference Sequence Database". National Center for Biotechnology Information. 14 March 2016. Retrieved 30 April 2016.
- ↑ Hsu, F., Kent, W.J.; Clawson, H.; Kuhn, R.M.; Diekhans, M.; Haussler, D. (2006). "The UCSC Known Genes". Bioinformatics. 22 (9): 1036–46. doi:10.1093/bioinformatics/btl048. PMID 16500937.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ a b Anders, S. (2010). "Counting reads in features with htseq-count". HTSeq 0.6.1p2 documentation. Retrieved 30 April 2016.
{{cite web}}: CS1 maint: uses authors parameter (link) - ↑ "DESeq". Bioconductor. Retrieved 30 April 2016.
- ↑ "DEXSeq". Bioconductor. Retrieved 30 April 2016.
- ↑ Anders, S.; Reyes, A.; Huber, W. (2012). "Detecting differential usage of exons from RNA-seq data". Genome Research. 22 (10): 2008–17. doi:10.1101/gr.133744.111. PMC 3460195. PMID 22722343.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Andrews, S. (8 March 2016). "FastQC". Babraham Bioinformatics. Babraham Institute. Retrieved 30 April 2016.
- ↑ "DNAA". SEQanswers. 19 December 2015. Retrieved 30 April 2016.
- ↑ a b "SAMtools". SourceForge. 12 September 2012. Retrieved 30 April 2016.
- ↑ "The bowtie aligner". Bowtie 1.1.2 Manual. Johns Hopkins University. 23 June 2015. Retrieved 30 April 2016.
- ↑ "USeq - Core Applications". SourceForge. Retrieved 30 April 2016.
- ↑ Kim, D.; Salzberg, S. (23 Februart 2016). "TopHat". Johns Hopkins University Center for Computational Biology. Retrieved 30 April 2016.
{{cite web}}: Check date values in:|date=(help)CS1 maint: multiple names: authors list (link) - ↑ Ameur, A.; Wetterbom, A.; Feuk, L.; Gyllensten, U. (2010). "Global and unbiased detection of splice junctions from RNA-seq data". Genome Biology. 11 (3): R34. doi:10.1186/gb-2010-11-3-r34. PMC 2864574. PMID 20236510.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ "PerM". Google Code Archive. Google. Retrieved 30 April 2016.
- ↑ "SOAPsplice". SOAP. Beijing Genomics Institute. 24 April 2013. Retrieved 30 April 2016.
- ↑ "GenomicFeatures". Bioconductor. Retrieved 30 April 2016.
- ↑ a b Robinson, M.D.; Oshlack, A. (2010). "A scaling normalization method for differential expression analysis of RNA-seq data". Genome Biology. 11 (3): R25. doi:10.1186/gb-2010-11-3-r25. PMC 2864565. PMID 20196867.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. (2010). "edgeR: A Bioconductor package for differential expression analysis of digital gene expression data". Bioinformatics. 26 (1): 139–40. doi:10.1093/bioinformatics/btp616. PMC 2796818. PMID 19910308.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ "GOseq". Bioconductor. Retrieved 30 April 2016.
- ↑ a b Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. (2010). "Gene ontology analysis for RNA-seq: accounting for selection bias". Genome Biology. 11 (2): R14. doi:10.1186/gb-2010-11-2-r14. PMC 2872874. PMID 20132535.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Li, H.; Lovci, M.T.; Kwon, Y.S.; Rosenfeld, M.G.; Fu, X.D.; Yeo, G.W. (2008). "Determination of tag density required for digital transcriptome analysis: Application to an androgen-sensitive prostate cancer model". Proceedings of the National Academy of Sciences of the United States of America. 105 (51): 20179–84. doi:10.1073/pnas.0807121105. PMC 2603435. PMID 19088194.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link)
Epigenetics
Epigenetics
Epigenetics is the science that studies inheritable traits not transmitted by plain sequence information. NGS can assess a particular non-standard epigenetics effect, which is the amount of methylation occurring on cytosines. This methylation is important biologically because it may influence the level of packing of chromatin and therefore affect efficiency of transcription in entire genomic areas. Cytosine methylation is reversible but inheritable somatically and germinally.
Genomic DNA can be treated with bisulfite[1], protocol that will transform only non-methylated cytosines into thymidines. Methylated cytosines are not affected, and will still be sequenced as such. A common NGS application in epigenetics is to align bisulfite-treated reads from a known organism on a reference genome, to assess the degree of methylation in particular areas. However, the complexity of the alignment will be higher, alongside with the reduced complexity of the reads (with several Cs being turned into Ts). Therefore, genomic DNA samples are sequenced both with and without bisulfite treatment, operation which allow to assess and normalize for the initial of reads aligning on specific regions.
Specific short read aligners exist for this task, just to name a few:
Protocols
Typical workflow
Chip-seq (Chromatin immunoprecipitation sequencing)
Chip-Sequencing is an effective technology that uses chromatin immunoprecipitation to DNA-protein or protein- protein interaction in the genome and it uses the more accurate, higher throughput method of sequencing.Even though ChIP-chip can be used to determine protein-DNA interaction, ChIP-seq is rapidly becoming the method of choice for the genome-wide localization of epigenetics, transcription regulation and post-transcription regulation.
The goal of ChIP-Seq method is to identify genome-wide binding patterns of a protein of interest. The major step involving ChIP-seq pipeline would be crosslinking, shearing, immunoprecipitation and sequencing. Formaldehyde is a widely used in DNA-protein crosslinking agent in ChIP methods. To perform ChIP, cells are treated with formaldehyde resulting in the covalent crosslinking of proteins to the DNA sequences which they are associated with.[2] Then, shearing of DNA is done either by sonication or by MNase digestion. The DNA is broken into pieces of about 0.2 to 1.0 kb in length. The fragmented DNA is purified using immunoprecipitation which is the process of binding antibody that are specific to the protein that is associated to DNA. The purified DNA-protein complexes are heated which separates protein from DNA. The fragmented DNAs are isolated and are sequenced by using next generation sequencing methods. Either 454, Solexa or Solid can be used to sequence based on the convenience.
File formats
Creating a dataset
Reference datasets
Viewing datasets
Comparing datasets
References
- ↑ Grunau, C.; Clark, S.J.; Rosenthal, A. (2001). "Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters". Nucleic Acids Research. 29 (13): e65. doi:10.1093/nar/29.13.e65. PMC 55789. PMID 11433041.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Barker, S.; Weinfeld, M.; Murray, D. (2005). "DNA–protein crosslinks: Their induction, repair, and biological consequences". Mutation Research. 589 (2): 111–135. doi:10.1016/j.mrrev.2004.11.003. PMC 15795165. PMID 11433041.
{{cite journal}}: Check|pmc=value (help)CS1 maint: multiple names: authors list (link)
Chromatin structure
Chromatin structure
Protocols
Typical workflow
File formats
Creating a dataset
Reference datasets
Viewing datasets
Comparing datasets
De novo assembly
De novo assembly
The generation of short reads by next generation sequencers has lead to an increased need to be able to assemble the vast amount of short reads that are generated. This is no trivial problem, as the sheer number of reads makes it near impossible to use, for example, the overlap layout consensus (OLC) approach that had been used with longer reads. Therefore, most of the available assemblers that can cope with typical data generated by Illumina use a de Bruijn graph based k-mer based approach.
A clear distinction has to be made by the size of the genome to be assembled.
- small (e.g. bacterial genomes: few Megabases)
- medium (e.g. lower plant genomes: several hundred Megabases)
- large (e.g. mammalian and plant genomes: Gigabases)
All de-novo assemblers will be able to cope with small genomes, and given decent sequencing libraries will produce relatively good results. Even for medium sized genomes, most de-novo assemblers mentioned here and many others will likely fare well and produce a decent assembly. That said, OLC based assemblers might take weeks to assemble a typical genome. Large genomes are still difficult to assemble when having only short reads (such as those provided by Illumina reads). Assembling such a genome with Illumina reads will probably will require using a machine that has about 256 GB and potentially even 512GB RAM, unless one is willing to use a small cluster (ABySS, Ray, Contrail), or invest into commercial software (CLCbio_Genomics_Workbench).
Typical workflow

A genome assembly project, whatever its size, can generally be divided into stages:
- Experiment design
- Sample collection
- Sample preparation
- Sequencing
- Pre-processing
- Assembly
- Post-assembly analysis
Experiment design
Like any project, a good de novo assembly starts with proper experimental design. Biological, experimental, technical and computational issues have to be considered:
- Biological issues: What is known about the genome?
- How big is it? Obviously, bigger genomes will require more material.
- How frequent, how long and how conserved are repeat copies? More repetitive genomes will possibly require longer reads or long distance mate-pairs to resolve structure.
- How AT rich/poor is it? Genomes which have a strong AT/GC imbalance (either way) are said to have low information content. In other words, spurious sequence similarities will be more frequent.
- Is is haploid, diploid, or polyploid? Currently genome assemblers deal best with haploid samples, and some provide a haploid assembly with annotated heterozygous sites. Polyploid genomes (e.g. plants) are still largely problematic.
- Experimental issues: What sample material is available?
- Is it possible to extract a lot of DNA? If you have only little material, you might have to amplify the sample (e.g. using MDA), thus introducing biases.
- Does that DNA come from a single cell, a clonal population, or a heterogeneous collection of cells? Diversity in the sample can create more or less noise, which different assemblers handle differently.
- Technical issues: What sequencing technologies to use?
- How much does each cost?
- What is the sequence quality? The greater the noise, the more coverage depth you will need to correct for errors.
- How long are the reads? The longer the reads, the more useful they will be to disambiguate repetitive sequence.
- Can paired reads be produced cost-effectively and reliably? If so, what is the fragment length? As with long reads, reliable long distance paired can help disambiguate repeats and scaffold the assembly.
- Can you use a hybrid approach? E.g. short and cheap reads mixed with long expensive ones.
- Computational issues: What software to run?
- How much memory do they require? This criteria can be final, because if a computer does not have enough memory, it will either crash, or slow down tremendously as it swaps data on and off the hard drive.
- How fast are they? This criteria is generally less stringent, since the assembly time is generally minor within a complete genome assembly and annotation project. However, some scale better than other.
- Do they require specific hardware? (e.g. large memory machine, or cluster of machines)
- How robust are they? Are they prone to crash? Are they well supported?
- How easy are they to install and run?
- Do they require a special protocol? Can they handle the chosen sequencing technology?
Some steps which are likely common to most assemblies:
- If it is within reason and would not tamper with the biology: Try to get DNA from haploid or at least mostly homozygous individuals.
- Make sure that all libraries are really ok quality-wise and that there is no major concern (e.g. use FastQC)
- For paired end data you might also want to estimate the insert size based on draft assemblies or assemblies which you have made already.
- Before submitting data to a de-novo assembler it might often be a good idea to clean the data, e.g. to trim away bad bases towards the end and/or to drop reads altogether. As low quality bases are more likely to contain errors, these might complicate the assembly process and might lead to a higher memory consumption. (More is not always better) That said, several general purpose short read assemblers such as SOAP de-novo and ALLPATHS-LG can perform read correction prior to assembly.
- Before running any large assembly, double and triple check the parameters you feed the assembler.
- Post assembly it is often advisable to check how well your read data really agrees with the assembly and if there are any problematic regions
- If you run de Bruijn graph based assemblies you will want to try different k-mer sizes. Whilst there is no rule of thumb for any individual assembly, smaller k-mers would lead to a more tangled graph if the reads were error free. Larger k-mer sizes would yield a less tangled graph, given error free reads. However, a lower k-mer size would likely be more resistant to sequencing errors. And a too large k might not yield enough edges in the graph and would therefore result in small contigs.
Data pre-processing
For a more detailed discussion, see the chapter dedicated to pre-processing.
Data pre-processing consists in filtering the data to remove errors, thus facilitating the work of the assembler. Although most assemblers have integrated error correction routines, filtering the reads will generally greatly reduce the time and memory overhead required for assembly, and probably improve results too.
Genome assembly
Genome assembly consists in taking a collection of sequencing reads, which are much shorter than the actual genome, and creating a genome sequence which is a likely source of all these fragments. What defines a likely genome depends generally on heuristics and the data available. Firstly, by parsimony, the genome must be as short as possible. One could take all the reads and simply produce the concatenation of all their sequences, but this wold not be parsimonious. Secondly, the genome must include as much of the input data as possible. Finally, the genome must satisfy as many of the experimental data as possibly. Typically, paired-end reads are expected to map onto the genome with a given respective orientation and a given distance from each other.
The output of an assembler is generally decomposed into contigs, or contiguous regions of the genome which are nearly completely resolved, and scaffolds, or sets of contigs which are approximately placed and oriented with respect to each other.
There are many assemblers available (See the Wikipedia page on sequence assembly for more details). Tutorials on how to use some of them are below.
Techniques for comparing assemblies
Once several genome assemblies are generated, they need to be evaluated.[1][2][3] Current methods include:
- N50 (length of contigs or scaffolds)[4]
- mapping of reads that were used to produce the assembly[5][6][7][8][9][10]
- identification and counting of highly conserved genes expected to be present based on evolution[11]
- mapping of transcripts to genome assemblies[12]
Post-assembly analysis
Once a genome has been obtained, a number of analyses are possible, if not necessary:
- Quality control
- Comparison to other assemblies
- Variant detection
- Annotation
Creating a dataset
Free Software
ABySS
ABySS is a de-novo assembler which can run on multiple nodes where it uses the message parsing interface (MPI) interface for communication. As ABySS distributes tasks, the amount of RAM needed per machine is smaller and thus Abyss is able to cope with large genomes. See here for a tutorial.
- Pros
- distributed interface a cluster can be used
- a large genome can be assembled with relatively little RAM per compute node. A human genome was assembled on 21 nodes having 16GB RAM each
- Cons
- relatively slow
Allpaths-LG
Allpath-LG is a novel assembler requiring specialized libraries. The authors of the software benchmarked ALLPATH-LG against SOAP-denovo and ALLPATH-LG reported superior performance. However it must be noted that they might not have used the SOAP-denovo gap filling module for one of the data set due to time constraints. This would probably have improved the SOAP assembly contiguous sequence length. In our own hand (usadellab) we have seen similar good N50 results[13] and also reported good N50 values for ALLPATHS-LG Arabidopsis assemblies. Similarly ALLPATHS-LG was named as well performing in the Assemblathon.
- Pros
- relatively fast runtime (slower than SOAP)
- good scaffold length (likely better than SOAP)
- can use long reads (e.g. PAC Bio) but only for small genomes
- Cons
- specially tailored libraries are necessary
- large genomes (mammalian size) need a lot of RAM. The publications estimates about 512GB would be sufficient though
- slower than SOAP
Euler SR USR
EULER is an assembler that includes an error correction module.
- Pros
- Has an error correction module
- Cons
MIRA
MIRA is a general purpose assembler that can integrate various platform data and perform true hybrid assemblies.
- Pros
- very well documented and many switches
- can combine different sequencing technologies
- likely relatively good quality data
- Cons
- Only partly multithreaded thus and due to the technology slow
- Probably not recommended to assemble larger genomes
Ray
Ray is a distributed scalable assembler tailored for bacterial genomes, metagenomes and virus genomes.
Tutorial available here
- Pros
- scalability (uses MPI)
- correctness
- usability
- well documented
- responsive mailing list
- can combine different sequencing technologies
- de Bruijn-based
- Cons
SOAP de novo
SOAPdenovo is an all purpose genome assembler. It was used to assemble the giant panda genome. See here for a tutorial.
- Pros
- SOAP de novo uses a medium amount of RAM
- SOAP de novo is relatively fast (probably the fastest free assembler)
- SOAP de novo contains a scaffolder and a read-corrector
- SOAP de novo is relatively modular (read-corrector, assembly, scaffold, gap-filler)
- SOAP de novo works well with very short reads[14]
- Cons
- potentially somewhat confusing way in which contigs are built.
- Relatively large amount of RAM needed, BGI states ca. 150GB (less than ALLPATHS though)
SPAdes
SPAdes is a single-cell genome assembler.
- Pros
- SPAdes works good with highly non-uniform coverage (e.g. after using Multiple Displacement Amplification)
- SPAdes uses medium ammount of RAM
- SPAdes is relatively fast
- SPAdes includes error correction software BayesHammer
- SPAdes have scaffolder (version 2.3+)
- Cons
- SPAdes is well tested only on bacterial genomes
- SPAdes works with Illumina reads only
Velvet
See here for a tutorial on creating an assembly with Velvet.
- Pros
- Easy to install, stable
- Easy to run
- Fast (multithreading)
- Can take in long and short reads, works with SOLiD colorspace reads
- Can use a reference genome to anchor reads which normally map to repetitive regions (Columbus module)
- Cons
- Velvet might need large amounts of RAM for large genomes, potentially > 512 GB for a human genome based if at all possible. This is based on an approximation formula derived by Simon Gladman[15] for smaller genomes -109635 + 18977*ReadSize + 86326*GenomeSize in MB + 233353*NumReads in million - 51092*Kmersize
Minia
Minia is a de Bruijn graph assembler optimized for very low memory usage.
- Pros
- Assembles very large genomes quickly on modest resources
- Easy to install, run
- Cons
- Illumina data only
- Does not perform any scaffolding
- Some steps are I/O-intensive, i.e. a local hard disk should be used rather than a network drive
Commercial
CLC cell
The CLC assembly cell is a commercial assembler released by CLC. It is based on a de Bruijn graph approach.
- Pros
- CLC uses very little RAM
- CLC is very fast
- CLC contains a scaffolder (version 4.0+)
- CLC can assemble data from most common sequencing platforms.
- Works on Linux, Mac and Windows.
- Cons
- CLC is not free
- CLC might be a bit more liberal in folding repeats based on our own plant data.
Newbler
Newbler is an assembler released by the Roche company.
- Pros
- Newbler has been used in many assembly projects
- Newbler seems to be able to produce good N50 values
- Newbler is often relatively precise
- Newbler can usually be obtained free of charge
- Cons
- Newbler is tailored to (mostly) 454 data. Since Ion Torrent PGM data has a similar error profile (predominance of miscalled homopolymer repeats), it may be a good choice there also. Whilst it can accommodate some limited amount of Illumina data as has been described by bioinformatician Lex Nederbragt[16], this is not possible for larger data sets. The fire ant genome[17] added ~40x Illumina data to ~15x 454 coverage in the form of "fake" 454 reads: first assembling the Illumina data using SOAPdenovo and then chopping the obtained contigs into overlapping 300bp reads, and finally inputting these fake 454 reads to Newbler alongside real 454 data.
- As Newbler at least partly uses the OLC approach large assemblies can take time
Decision Helper
This is based both on personal experience as well as on published studies. Please note however that genomes are different and software packages are constantly evolving.
An Assemblathon challenge which uses a synthetic diploid genome assembly was reported on by Nature to call SOAP de novo, Abyss and ALLPATHS-LG the winners.[18]
However a talk on the Assemblethon website names SOAP de novo, sanger-sga and ALLPATHS-LG to be consistently amongst the best performers for this synthetic genome.[19]
I want to assemble:
- Mostly 454 or Ion Torrent data
- small Genome =>MIRA, Newbler
- all others use Newbler
- Mixed data (454 and Illumina)
- small genome => MIRA, but try other ones as well
- medium genome => no clear recommendation
- large genome, assemble Illumina data with ALLPATHS-LG and SOAP, add in other reads or use them for scaffolding
- Mostly Illumina (or Colorspace)
- small genome => MIRA, velvet
- medium genome => no clear recommendation
- large genome, assemble Illumina data with ALLPATHS-LG and SOAP, add in other reads or use them for scaffolding
(For large genomes this is based on the fact that not many assemblers can deal with large genomes, and based on the assemblathon outcome. For 454 data this is based on Newbler's good general performance, and MIRA's different outputs, its versatility and the theoretical consideration that de Bruijn based approaches might fare worse)
Post assembly you might want to try the SEQuel software to improve the assembly quality.
I want to start a large genome project for the least cost
- Use Illumina reads with ALLPATHS-LG specification (i.e. overlapping), the reads will work in e.g. SOAP de novo as well
(This recommendation is based on the Assemblathon outcome, the original ALLPATHS publication[20] as well as a publication that used ALLPATHS for the assembly of Arabidopsis genomes.[13]
Each software has its particular strength, if you have specific requirement, the result from Assemblathon will guide you. Another comparison site GAGE has also released its comparison.[2] Also there exists QUAST tool for assessing genome assembly quality.
Case study
Further Reading Material
- Background
- Genome Sequence Assembly Primer
- Paszkiewicz and Studholme, 2010 Some general background
- Nagarajan and Pop, 2010
- Imeldfort and Edwards, 2009 Sequencing of plant genomes
- Pop 2009
- Original publications
- Simpson et al., 2009 ABySS
- Zerbino and Birney, 2008 Velvet
- Gnerre et al., 2011 ALLPATHS-LG
- Li et al., 2010 SOAP denovo
- Chevreaux et al., 2004 MIRAest
- Chaisson et al., 2009 EULER-USR
- The CLC Assembly Cell Whitepaper includes a comparison with ABySS for a human genome and with velvet for a bacterial genome
- Comparisons
- Ye et al., 2011 Comparison of Sanger/PCAP; 454/Roche and Illumina/SOAP assemblies. Illumina/SOAP had lower substitution, deletion and insertion rates but lower contig and scaffold N50 sizes than 454/Newbler.
- Paszkiewicz et al., 2010 General review about short read assemblers
- Zhang et al., 2011 In depth comparison of different genome assemblers on simulated Illumina read dat. Unfortunately only up to medium genomes were tested. For eukaryotic genomes and short reads Soap denovo is suggested for longer reads ALLPATHS-LG.
- Chapman JA et al. 2011 introduce the new assembler Meraculous gathered literature data on the assembly of E. coli K12 MG1655 for Allpaths 2, Soapdenovo, Velvet, Euler-SR, Euler, Edena, AbySS and SSAKE. Allpaths2 had by far the largest Contig and Scaffold N50 and was apart from Meraculous the only misassembly free. Meraculous was shown to even contain no errors.
- Liu et al., 2011 benchmark their new assembler PASHA against SOAP de novo (v 1.04), velvet (1.0.17) and ABySS (1.2.1) using three bacterial data sets. Whilst PASHA usually the largest NG50 and NG80 (N50 and N80 calculated with the true genome sizes) SOAP de novo produced the highest number of contigs and soemtimes worse NG50 and NG80. However for one dataset SOAP denovo showed the best genome coverage.
- The Assemblathon comparing de novo genome assemblies of many different teams based on a synthetic genome. The Assemblathon 1 competition is now published in Genome Research by Earl et al.[1]
Reference datasets
ENA
See here for more information.
The European Nucleotide Archive (ENA), has a three-tiered data architecture.It consolidates information from:
- EMBL-Bank.
- the European Trace Archive:containing raw data from electrophoresis-based sequencing machines.
- the Sequence Read Archive: containing raw data from next-generation sequencing platforms.
SRA
See SRA for more information.
The Sequence Read Archive (SRA) is:
- the Primary archival repository for next generation sequencing reads and alignments (BAM)
- Expanding to manage other high-throughput data including sequence variations (VCF)
- Will shorty also accept capillary sequencing reads
- Globally comprehensive through INSDC data exchange with NCBI and DDBJ
- Part of European Nucleotide Archive (ENA)
- Data owned by submitter and complement to publication
- Data expected to be made public and freely available; no access/use restrictions permitted
- Pre-publication confidentiality supported
- Controlled access data submitted to EGA
- Active in the development of sequence data storage and compression algorithms/technologies
SRA Metadata Model
- Study: sequencing study description
- Sample: sequenced sample description
- Experiment/Run: primary read and alignment data
- Analysis: secondary alignment and variation data
- Project: groups studies together
- EGA DAC: Data Access Committee
- EGA Policy: Data Access Policy
- EGA Dataset: Dataset controlled by Policy and DAC
NCBI
Viewing datasets
ENSEMBL
UCSC
Tablet
IGV
IGV is the Integrative Genomics Viewer developed by NCBI, the National Center for Biotechnology Information. IGV allows for easy navigation of large-scale genomic datasets, and supports the integration of genomic data types such as aligned sequence reads, mutations, copy number, interfering RNA screens, gene expression, methylation, and genomic annotations. Users can amplify specific areas down to individual base-pairs, and more generally scroll through an entire genome. It can be used to visualize and share whole genomes/reference genomes, alignments, variants, and regions of interest as well as filter, sort, and group genomic data.
Comparing datasets
Whole genome alignments
References
- ↑ a b Earl, D.; Bradnam, K.; St. John, J.; et al. (2011). "Assemblathon 1: A competitive assessment of de novo short read assembly methods". Genome Research. 21 (12): 2224–41. doi:10.1101/gr.126599.111. PMC 3227110. PMID 21926179.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ a b Salzberg, S.L.; Phillippy, A.M.; Zimin, A.; et al. (2012). "GAGE: A critical evaluation of genome assemblies and assembly algorithms". Genome Research. 22 (3): 557–67. doi:10.1101/gr.131383.111. PMC 3290791. PMID 22147368.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Bradnam, K.R.; Fass, J.N.; Alexandrov, A.; et al. (2013). "Assemblathon 2: Evaluating de novo methods of genome assembly in three vertebrate species". GigaScience. 2 (1): 10. doi:10.1186/2047-217X-2-10. PMC 3844414. PMID 23870653.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Mäkinen, V.; Salmela, L.; Ylinen, J. (2012). "Normalized N50 assembly metric using gap-restricted co-linear chaining". BMC Bioinformatics. 13: 255. doi:10.1186/1471-2105-13-255. PMC 3556137. PMID 23031320.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Ghodsi, M.; Hill, C.M.; Astrovskaya, I.; et al. (2013). "De novo likelihood-based measures for comparing genome assemblies". BMC Research Notes. 6: 334. doi:10.1186/1756-0500-6-334. PMC 3765854. PMID 23965294.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Hunt, M.; Kikuchi, T.; Sanders, M.; et al. (2013). "REAPR: A universal tool for genome assembly evaluation". Genome Biology. 14 (5): R47. doi:10.1186/gb-2013-14-5-r47. PMC 3798757. PMID 23710727.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Phillippy, A.M.; Schatz, M.C.; Pop, M. (2008). "Genome assembly forensics: Finding the elusive mis-assembly". Genome Biology. 9 (3): R55. doi:10.1186/gb-2008-9-3-r55. PMC 2397507. PMID 18341692.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Rahman, A.; Pachter, L. (2013). "CGAL: Computing genome assembly likelihoods". Genome Biology. 14 (1): R8. doi:10.1186/gb-2013-14-1-r8. PMC 3663106. PMID 23360652.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Vezzi, F.; Narzisi, G.; Mishra, B. (2012). "Reevaluating assembly evaluations with feature response curves: GAGE and Assemblathons". PLoS One. 7 (12): e52210. doi:10.1371/journal.pone.0052210. PMC 3532452. PMID 23284938.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Howison, M.; Zapata, F.; Dunn, C.W. (2013). "Toward a statistically explicit understanding of de novo sequence assembly". Bioinformatics. 29 (23): 2959–63. doi:10.1093/bioinformatics/btt525. PMID 24021385.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Parra, G.; Bradnam, K.; Korf, I. (2007). "CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes". Bioinformatics. 23 (9): 1061–7. doi:10.1093/bioinformatics/btm071. PMID 17332020.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Ryan, J.F. (7 February 2014). "Baa.pl: A tool to evaluate de novo genome assemblies with RNA transcripts". Cornell University Library. Retrieved 4 May 2016.
- ↑ a b Schneeberger, K.; Ossowski, S.; Ott, F.; et al. (2011). "Reference-guided assembly of four diverse Arabidopsis thaliana genomes". PNAS. 108 (25): 10249–54. doi:10.1073/pnas.1107739108. PMC 3121819. PMID 21646520.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Zhang, W.; Chen, J.; Yang, Y.; et al. (2011). "A practical comparison of de novo genome assembly software tools for next-generation sequencing technologies". 6 (3): e17915. doi:10.1371/journal.pone.0017915. PMC 3056720. PMID 21423806.
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|author=(help); Text "journal PLoS One" ignored (help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Gladman, S. (23 July 2009). "(Velvet-users) Velvetg running time". Velvet-users mailing list. The European Bioinformatics Institute. Retrieved 4 May 2016.
- ↑ Nederbragt, L. (21 January 2011). "Newbler input II: Sequencing reads from other platforms". An assembly of reads, contigs and scaffolds. Retrieved 4 May 2016.
- ↑ Wurm, Y.; Wang, J.; Riba-Grognuz, O.; et al. (2010). "The genome of the fire ant Solenopsis invicta". PNAS. 108 (14): 5679–84. doi:10.1073/pnas.1009690108. PMC 3078418. PMID 21282665.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link) - ↑ Hayden, E.C. (2011). "Genome builders face the competition". Nature. 471 (7339): 425. doi:10.1038/471425a. PMID 21430748.
- ↑ "Assemblathon 1 results". assemblathon.org. University of California - Davis. 1 June 2011. Retrieved 4 May 2016.
- ↑ Gnerre, S.; Maccallum, I.; Przybylski, D. (2011). "High-quality draft assemblies of mammalian genomes from massively parallel sequence data". Proceedings of the National Academy of Sciences of the United States of America. 108 (4): 1513–8. doi:10.1073/pnas.1017351108. PMC 3029755. PMID 21187386.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: multiple names: authors list (link)
Velvet
Velvet practical: Part 1
This practical will cover the following items:
- Compile Velvet
- Single end
- K-mer length
- Coverage cut-offs
- Whole genome sequence as input???
Prepare the environment
First make sure that you are in your home directory by typing:
cd
and making absolutely sure you're there by typing:
pwd
Now create sub-directories for this and the two other velvet practicals. All these directories will be made as sub-directories of a directory for the whole course called NGS. For this you can use the following commands:
mkdir -p NGS/velvet/{part1,part2,part3}
# The -p tells mkdir (make directory) not to worry if a parent directory is missing. # That is, if a sub-directory cannot be made because its parent directory does not exist, # just make the parent directory first rather than reporting an error. # The “one at a time” approach would be: mkdir NGS mkdir NGS/velvet mkdir NGS/velvet/part1 mkdir NGS/velvet/part2 mkdir NGS/velvet/part3
After creating the directories, examine the structure and move into the directory ready for the first velvet exercise by typing:
ls -R NGS;
cd NGS/velvet/part1; pwd;
Downloading and Compile Velvet
You can find the latest version of velvet at:
http://www.ebi.ac.uk/~zerbino/velvet/
You could go to this URL and download the latest velvet version, or equivalently, you could type the following, which will download, unpack, inspect, compile and execute your locally compiled version of velvet:
cd ~/NGS/velvet/part1; pwd;
cp ~/NGS/Data/velvet_1.2.07.tgz . ;
tar xzf ~/NGS/Data/velvet_1.2.07.tgz;
ls -R; cd velvet_1.2.07;
make velveth velvetg;
./velveth'''
Take a look at the executables you have created. They will be displayed as green by the command:
ls --color=always;
The switch --color, instructs that files be coloured according to their type. This is often the default. Here we are just being cautious. The =always is included as colouring is usually suppressed in scripts. If you run this exercise using the script provided, just --color would not be enough. --color=always insists on file colouring even from a script.
Have a look of the output the command produces and you will see the following parameters passed into the compiler:
“MAXKMERLENGTH=31” and “CATEGORIES=2”
This indicates that the default compilation was set for De Bruijn graph KMERs of maximum size 31 and to allow a maximum of just 2 read categories. You can override these, and other, default configuration choices using command line parameters. Assume, you want to run velvet with a KMER length of 41 using 3 categories, velvet needs to be recompiled to enable this functionality by typing:
make clean; make velveth velvetg MAXKMERLENGTH=41 CATEGORIES=3;
./velveth
velvet can also be used to process SOLID colour space data. To do this you need a further make parameter. With the following command clean away your last compilation and try the following parameters:
make clean; make MAXKMERLENGTH=41 CATEGORIES=3 color
./velveth_de
For a further description of velvet compile and runtime parameters please see the velvet Manual: https://github.com/dzerbino/velvet/wiki/Manual
Single ended read assembly
The data you will examine is from Staphylococcus aureus USA300 which has a genome of around 3MB. The reads are Illumina and are unpaired, also known as single-end library. Even though you have carefully installed velvet in your own workspace, we will use a pre-installed version. The data needed for this section can be obtained from the Sequence Read Archive (SRA). For the following example use the run data SRR022825 and SRR022823 from the SRA Sample SRS004748. The SRA experiment could be viewed by setting your browser to the URL: http://www.ebi.ac.uk/ena/data/view/SRS004748
The following exercise focuses on velvet using single-end reads, how the available parameters effect an assembly and how to measure and compare the changes. To begin with, first move back to the directory you prepared for this exercise, create a new folder with a suitable name for this part and move into it. The command to download the file from the internet would be:
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR022/SRR022825/SRR022825.fastq.gz
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR022/SRR022823/SRR022823.fastq.gz
or if you had the files installed locally, just create soft links to the files. Continue by copying (or typing):
<syntax highlight="text> cd ~/NGS/velvet/part1 mkdir SRS004748 cd SRS004748 pwd ln -s ~/NGS/Data/SRR022825.fastq.gz . ln -s ~/NGS/Data/SRR022823.fastq.gz . ls -l </syntaxhighlight>
You are ready to process your data with velvet, which is a compressed fastq file. Velvet has two main components: velveth -used to construct, from raw read data, a dataset organised in the fashion expected by the second component, velvetg. velvetg -the core of velvet where the de Bruijn graph assembly is built and manipulated. You can always get further information about the usage of both velvet programs by typing velvetg or velveth in your terminal.
Now run velveth for the reads in SRR022825.fastq.gz and SRR022823.fastq.gz using the following options:
- A de Bruijn graph k-mer of 25
- An output directory called run_25
velveth run_25 25 -fastq.gz -short SRR022825.fastq.gz SRR022823.fastq.gz
velveth talks to itself for a while and ends with some files in the output directory. Move into the output directory run_25 and take a look around at what velveth had done so far. The UNIX command less allows you to look at output files (press q for quit). Just in case you still need a hint:
cd run_25;
ls -l;
head Sequences;
Now move one directory level up and run velvetg on your output directory, with the commands:
cd ..
time velvetg run_25
Move back into your results directory to examine the effects of velvetg:
cd run_25; ls -l;
FOR YOU: once you run the command above:
- Q1: What extra files do you see in the folder run_25?
- Q2:What do you suppose they might represent?
- Q3: In the Log file in run_25, what is the N50?
N50 statistic: Broadly, it is the median (not average) of a sorted data set using the length of a set of sequences. Usually it is the length of the contig whose length. When added to the length of all longer contigs, makes a total greater that half the sum of the lengths of all contigs. Easy, but messy – a more formal definition can be found here: http://www.broadinstitute.org/crd/wiki/index.php/N50
Backup the contigs.fa file and calculate the N50 (and the N25,N75) value with the command:
cp contigs.fa contigs.fa.0
YOU now try:
gnx -min 100 -nx 25,50,75 contigs.fa
- Q4. Does the value of N50 agree with the value stored in the Log file?
- Q5. If not, why do you think this might be?
In order to improve our results, take a closer look at the standard options of velvetg by typing 'velvetg' without parameters. For the moment focus on the two options -cov_cutoff and -exp_cov. Clearly -cov_cutoff will allow you to exclude contigs for which the kmer coverage is low, implying unacceptably poor quality. The -exp_cov switch is used to give velvetg an idea of the coverage to expect. If the expected coverage of any contig is substantially in excess of the suggested expected value, maybe this would indicate a repeat. For further details of how to choose the parameters, go to 'Choice of a coverage cutoff': http://wiki.github.com/dzerbino/velvet/
Briefly, the Kmer coverage (and much more information) for each contig is stored in the file stats.txt and can be used with R to visualize the coverage distribution. Take a look at the stats.txt file, start R, load and visualize the data using the following commands:
R
library(plotrix)
data <- read.table("stats.txt", header=TRUE)
x11()
weighted.hist(data$short1_cov, data$lgth, breaks=0:50)
A weighted histogram is a better way of visualizing the coverage information, because of noise (lot of very short contigs). You can see an example output below:

After choosing the expected coverage and the coverage cut-off, you can exit R by typing:
q()
n
The weighted histogram suggests to me that the expected coverage is around 14 and that everything below 6 is likely to be noise.
Some coverage is also represented at around 20, 30 and greater 50, which might be contamination or repeats (depending on the dataset), but at the moment this should not worry you.
To see the improvements, rerun velvetg first with -cov_cutoff 6 and after checking the N50 use only / add -exp_cov 14 to the command line option.
Also keep a copy of the contigs file for comparison:
cd ~/NGS/velvet/part1/SRS004748
time velvetg run_25 -cov_cutoff 6
# Make a copy of the run
cp run_25/contigs.fa run_25/contigs.fa.1
time velvetg run_25 -exp_cov 14
cp run_25/contigs.fa run_25/contigs.fa.2
time velvetg run_25 -cov_cutoff 6 -exp_cov 14
cp run_25/contigs.fa run_25/contigs.fa.3
- Q6. What is the N50 with no parameter:
- Q7. What is the N50 with -cov_cutoff 6:
- Q8. What is the N50 with -exp_cov 14:
- Q9. What is the N50 with -cov_cutoff 6 -exp_cov 14:
- Q10Did you notice a variation in the time velvetg took to run? If so, can you explain why that might be?
You were running velvetg with the given -exp_cov and -cov_cutoff parameters. Now try to experiment using different cut-offs, expected parameters and also explore other settings (e.g. -max_coverage, -max_branch_length, -unused_reads, -amos_file, -read_trkg or see velvetg help menu).
In particular, look at the -amos_file parameter which instructs velvetg to create a version of the assembly that can be processed and viewed with a program called AMOS Hawkeye. Another program, called tablet, can also understand and display the AMOS file. For now, we will take a look at tablet.
Run velvetg with just -cov_cutoff 6 but requesting an amos file:
velvetg run_25 -cov_cutoff 6 -amos_file yes
Now take a swift look at the assembly with tablet:
tablet run_25/velvet_asm.afg &
velvet_asm.afg being the file velvetg made is response to the inclusion of the -amos_file yes switch. tablet will take quite a bit of memory and you can safely ignore the complaints from tablet. Once the file has loaded, select one of the longer contigs. Note the lack of Read information and the one dimensional nature of the contig display. Close down tablet when you have seen enough. Now rerun velvetg adding the additional parameter -exp_cov 7 with no need to save any files as the amos file needs to be changed. Now rerun velvetg adding the additional parameter -exp_cov 14 with no need to save any files as the AMOS file needs to be changed.
velvetg run_25 -cov_cutoff 6 -exp_cov 14 -amos_file yes
Again, view the amos file with tablet:
tablet run_25/velvet_asm.afg &
Select a longish contig as before and explore the display options before close down table. FOR YOU:
- Q11. Why do you think there was read information only for the second use of tablet?
- Q12. You may have noticed velvetg took a little longer with -exp_cov 14 on. Does this make sense ?
If you want to explore the behaviour of velvet even further, you could experiment with the following.
Reduce the sequence coverage by only choosing one input file for the assembly e.g. SRR022825.fastq.gz
Increase the sequence coverage by downloading further files from the same ENA sample SRS004748 (http://www.ebi.ac.uk/ena/data/view/SRS004748).
FOR YOU: How did the N50 change by
- Q13. reducing the coverage?
- Q14. increasing the coverage?
