Three Dimensional Electron Microscopy/Printable version
| This is the print version of Three Dimensional Electron Microscopy You won't see this message or any elements not part of the book's content when you print or preview this page. |
The current, editable version of this book is available in Wikibooks, the open-content textbooks collection, at
https://en.wikibooks.org/wiki/Three_Dimensional_Electron_Microscopy
What is 3DEM?
What is 3DEM?
[edit | edit source]

Cryogenic electron microscopy, often abbreviated as ‘cryo-EM’ has evolved to encompass a wide range of experimental methods. Cryo-EM is increasingly becoming a mainstream technology for studying cells, viruses, and protein structures at molecular resolution[1]. Images are produced using a electron microscope, using electrons as radiation, emitted by a source that is housed under a high vacuum, and then pushed down the microscope column at accelerating voltages in the range of 80-300 kV[1]. A very large difference in electron microscopy compared to optical microscopy is the resolving power of the two methods, with electron microscopy having a much high resolving power. The resolving power of a microscope is directly related to the wavelength of the irradiation used to form an image. The electron irradiation used causes extensive damage to the biological sample. One-way to reduce the electron induced sample damage is to perform the technique in cryogenic temperatures. Freezing aqueous solutions in cryogen, such as liquid ethane cooled by liquid nitrogen is a method commonly used to prepare specimens for cryo-EM applications[1]. This method has proved to reduce the effects of radiation damage in a large way, which in turn lets researchers use higher doses of electrons to increase the signal to noise ratio, because of less radiation damage. The commonly used variant of cryo-EM is single particle analysis. Using this technique data from a large number of 2D projection images such as identical copies of protein complexes in different orientations are combined to generate a 3D reconstruction of the structure [1].
Particle Picking
[edit | edit source]Particle picking attempts to correctly the position of particles, and differentiate between the particles and any contamination in an image [2]. Particles may be chosen manually or through the use of an automated algorithm. The automated algorithm strives to separate the “positive class” of particles from the “negative class” of contaminants or noise[3]. Automated algorithms may be prone to a type 1 or type 2 error, which falsely identify positives or negatives. .
Contrast Transfer Function
[edit | edit source]The contrast transfer function (CTF) is a distortion of the image data collected due to flawed optical properties in a transmission electron microscope[4]. There are two cause of the CTF and both are related to the lens system in electron microscopy. The first cause is spherical aberrations that cause many focal points, which blurs the image. The second cause of the CTF is the under focus used in electron microscopy. To obtain a high resolution 3D image, the CTF must be corrected.
Particle Extraction and Stack Creation
[edit | edit source]A stack is a collection of similar images of the same structure. Individual images can be obtained by removing unnecessary information to increase processing speed[5]. Particle boxing cuts out the image 150% the particle’s pixel size. Particle contrast differentiates between particles on a negative stain and on vitreous ice, inverting when the particles are black against a light background. CTF correction alternates the particles between the data and its inverse to correct the particles.
Particle Alignment
[edit | edit source]In particle alignment images of a particle are shifted and rotated in many different directions to attempt to align the particles. They must be shifted and rotated to align together to obtain an average image. The goal when averaging is to maximize the cross correlation of the images[6]. Particle alignment can either be classified as reference based or reference free. Depending on the classification, different types of automated packages may be used for alignment e.g. Xmipp Maxium Likelihood Alignment, Spider Reference Liklihood, and EMAN Refine 2D Refrence-free Alignment[7].
Initial Model Creation
[edit | edit source]Multiple 2D images are required to obtain the structure of a 3D image. Particles are chosen and organized by structural characteristics including common lines, shapes, and tilts[8]. The particles are clustered with other structurally similar particles, and then averaged together [2]. Reconstruction of a 3D model is reliant on the particle orientation, as stated by the central projection theorem[1].
3D Reconstruction
[edit | edit source]After getting an initial 3D structure by using one of a number of methods, such as random conical tilt or angular reconstitution, the obtained map is used as a 3D reference to refine the single particle orientation parameters (two for position and three for orientation). The structure can then be iteratively improved, alternating position and orientation determination with 3D reconstruction. The FSC Fourier shell correlation) and other resolution criteria can be used to follow the progress of the structure refinement, which is terminated when there is no further improvement. Different program such as IMAGIC, SPIDER, FREALIGN can be used for this purpose[9].
References
[edit | edit source]- ↑ a b c d e Milne, Jacqueline L, Mario J Borgnia, and Alberto Bartesaghi. "Cryo-electron microscopy – a primer for the non-microscopist." Febs Journal.280 (2012): n. page. Print.
Invalid
<ref>tag; name "electron" defined multiple times with different content - ↑ a b Voss, Neil. "Particle Picking." Roosevelt University, Schaumburg. 13 Sept. 2013. Lecture.
- ↑ Langlois, Robert, and Joachim Frank. "A Clarification of the Terms Used in Comparing Semi-automated Particle Selection Algorithms in Cryo-EM." Journal of Structural Biology 175.3 (2011): 348-52. Science Direct. Howard Hughes Medical Institute. Web. 3 Dec. 2013.
- ↑ Voss, Neil. "Contrast Transfer Function." Roosevelt University, Schaumburg. 27 Sept. 2013. Lecture.
- ↑ Voss, Neil. "Particle Boxing." Roosevelt University, Schaumburg. 27 Sept. 2013. Lecture.
- ↑ Sigworth, F.J. "A Maximum-Likelihood Approach to Single-Particle Image Refinement." Journal of Structural Biology.122 (1998): n. page. Print.
- ↑ Voss, Neil. "Particle Alignment." Roosevelt University, Schaumburg. 4 Oct. 2013. Lecture.
- ↑ Voss, Neil. "Initial Model Problem." Roosevelt University, Schaumburg. 8 Nov. 2013. Lecture.
- ↑ Voss, Neil. "3D Reconstruction." Roosevelt University, Schaumburg. 25 Oct. 2013. Lecture.
Electron microscopes
What is an Electron Microscope?
[edit | edit source]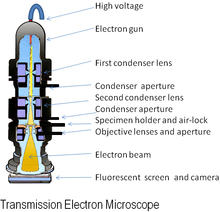
An electron microscope (EM) is an imaging instrument that uses electrons to see a sample instead of light which is used in the traditional light microscope. In general, an electron microscope works by applying a beam of electrons to a very thinly sliced or diluted sample. The electrons will either bounce off or pass through the sample and an image will be collected depending on the type of microscopy being utilized.
The resolution capability of an electron microscope is much greater than that of a light microscope, generally obtaining a magnification of 100,000X which is 50,000X greater than a traditional light microscope. The resolution difference can be attributed to the energy source of the microscopes as well as the methods of detection by instruments far more sensitive than the human eye. Light microscopes are capable of resolving about 200 nm in size. Electron microscopes in have the ability to resolve approximately 2 Å. There have been reports of electron microscopes reaching a resolution of 0.5 Å[1].
An Electron Microscope vs. a Traditional Light Microscope
[edit | edit source]A traditional light microscope and an electron microscope operate on similar principles. Both microscopes contain an energy source, a condenser lens, a specimen holder, an objective lens, and a projector lens. In both cases, energy is directed at a sample and an image is produced. A major differing feature of each microscope is the energy source. In an electron microscope, electrons are emitted from an electron gun, while in the light microscope the energy is generated by a light bulb. Another important difference between the two microscopes is the composition of the lens. The lens of a light microscope are made of glass and are typically spherical. In contrast, an electron microscope’s lenses are electromagnetic and electrostatic. The electromagnetic lens mainly consists of coiled copper wires. In the optical microscope light travels from the source, is split up, and then is refocused to be viewed by the human eye. In an electron microscope the electrons are passed through or bounced off of the sample and are ultimately imaged on film, a fluorescent screen or collected by an electron detector.
A unique feature of the electron microscope is the sample holder and environmental condition of the sample chamber. The sample grid, either suspended in vitreous ice or negatively stained, is placed onto the electron microscope holder. The holder is then inserted into a closed chamber, where it can be rotated at varying degrees to be imaged at specific, user selected, angles. The environment of the microscope chamber is maintained under vacuum to facilitate the directed travel of the electron beam.
Types of Microscopy
[edit | edit source]There are several types of microscopy where an electron microscope is used. The various electron microscopy techniques are Scanning Electron Microscopy (SEM), Transmission Electron Microscopy (TEM), Reflection Electron Microscopy (REM), and Scanning Transmition Microscopy (STEM). The most commonly used techniques are SEM and TEM.
Transmission Electron Microscopy works by directing the flow of electrons at a sample. TEM requires a very thin sample specimen that can endure high energy electrons, as the electrons are transmitted through the sample. As the electrons of the beam interact with the sample, an image is formed. In essence, the “shadow” of the image, created in the places were the electrons interacted with the sample, is projected onto the detector.
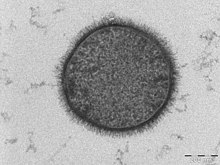
During SEM, an electron beam scans the surface of the sample and the electrons or X rays that are emitted from the surface are detected. The signals produced are used to derive information about the sample's surface topography. Various types of energy can be used, but the most common is secondary electrons excited by the initial interaction with the electron beam. SEM details the surface of a sample, while TEM details the entire sample.

History
[edit | edit source]The first model of an electron microscope was built by Ernst Ruska and Max Knoll in 1931[2]. This initial model was only capable of magnification at 400x. Two years later, an electron microscope capable of magnifications greater than the traditional microscope was developed. In 1938 Max Knoll created the first scanning electron microscope which was refined by Manfred Von Ardenne to give better resolution.
Current Applications
[edit | edit source]The Electron Microscope in currently used in numerous applications such as rapid medical diagnosis[3], pharmaceutical nano-scale systems[4] and failure mode analysis of components, but is most commonly used in the imaging of molecules. The techniques were most recently used in the imaging of double stranded DNA[5].
References
[edit | edit source]- ↑ Electron Microscope Breaks Half-Angstrom Barrier. Physics World. [Online]. Accessed 02 Dec. 13. Available from: http://physicsworld.com/cws/article/news/2007/sep/17/electron-microscope-breaks-half-angstrom-barrier
- ↑ History of the Electron Microscope in Cell Biology.Barry R Masters, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA. Online. Accessed 04 Dec 2013. Available from: http://www.fen.bilkent.edu.tr/~physics/news/masters/ELS_HistoryEM.pdfhttp://www.fen.bilkent.edu.tr/~physics/news/masters/ELS_HistoryEM.pdf
- ↑ Electron Microscopy for Rapid Diagnosis of Emerging Infectious Agents.P Hazelton and H Gelderblom. Center for Disease Control. [Online]. Accessed 04 Dec 2013. Available from: http://wwwnc.cdc.gov/eid/article/9/3/02-0327_article.htm
- ↑ Electron Microscopy of Pharmaceutical systems. V Klang, C Valenta, N Matsko, Micron, 2013, 44, pp 45-74.
- ↑ Direct Imaging of DNA Fibers: The Visage of Double Helix. F Gentile, M Moretti, T Limongi, A Falqui, G Bertoni, A Scarpellini, S Santoriello, L Maragliano, R Zaccaria, and E di Fabrizio. Nano Lett., 2012, 12 (12), pp 6453–6458.
Particle Picking
What is Particle Picking
[edit | edit source]w:Particle picking takes the image obtained from the electron microscopy and its goal it to obtain individual particles from the image. For some particles it works best with certain particle picking programs since many particles have different orientations. Finding individual particles could be done in four ways. The four ways to find individual particles:
- Manual Selection
- Template matching
- Mathematical Function
- Machine Learning
Manual Selection
[edit | edit source]
Manual picking is a technique of particle picking that requires the user to manually select the particles from the micrograph. Manual picking is a time consuming technique, which makes it the less preferable over other methods of particle selection. In order to obtain better quality, the user need avoid the particles that are close to the edges, the particles that overlap each other, and the contaminated particles.
Template Matching
[edit | edit source]w:Template matching is a digital technique that is used for object classification. The template matching technique can be used to find particular parts in an image by comparing it to a similar template image. It requires a sample patch image that is used to identify most similar area in the source image [1]. Templates are often used to identify numbers, characters, and any kind of small areas in images. The patch is compared by sliding it on one pixel at a time [2]. Consequently, it calculates an index number that represents how similar the template matches the image in that particular position[1]. The similarity to the template image can be calculated using the correlation coefficient.
Equation to find index number:
Mathematical Function
[edit | edit source]The use of mathematical function helps form templates by using the w:Difference of Gaussian (abbreviated to DoG). DoG picker sorts particle by its size using the radius of a particle and it works along as a reference-free particle picker[3]. The Mathematical function works best with particles that are symmetrical in shape, however not all particles are symmetrical and many are asymmetrical. Unlike manual selection, there are some errors that may occur when using DoG picker. As what was said before some particles are asymmetrical and the DoG picker will have difficulties picking those particles. [3].
Machine Learning
[edit | edit source]Machine learning technique provides automatic particle selection from electron micrograph. Through automatic particle picking, a large number of particles can be selected in a short period of time without the need of extensive manual intervention. The machine learning method can utilize an algorithm to discriminate particles from non-particles [4] The algorithm can be corrected during picking in the training phase to minimize the number of false positives [5] [4]. The training phase has to be semi-supervised to allow the user for algorithm correction to identify the particles selected incorrectly [4]. The method eliminates the need for initial reference volume as it learns the particles of interest from the user.
Reference
[edit | edit source]- ↑ a b Template Matching." Template Matching — OpenCV 2.4.7.0 documentation. N.p., n.d. Web. 22 Nov. 2013
- ↑ Jan Latecki, Longin . "Template Matching." Template Matching. N.p., n.d. Web. 22 Nov. 2013.
- ↑ a b Voss, N.R., C.K. Yoshioka, M. Radermacher, C.S. Potter, and B. Carragher. "DoG Picker and TiltPicker: Software Tools to Facilitate Particle Selection in Single Particle Electron Microscopy." Journal of Structural Biology 166.2 (2009): 205-13. Print.
- ↑ a b c Sorzano C.O.S, Recarte E, Alcorio M, Bibao-Castro J.R., San-Martin C, Marabini R, Carazo J.M., (2009). Automatic particle selection from electron micrograph using machine learning techniques. J Structural Biol. 167 (3), pp.252-260.
- ↑ Zhu Y, Carragher B, Glaeser RM, Fellmann D, Bajaj C, Bern M, Mouche F, de Haas F, Hall RJ, Kriegman DJ, Ludtke SJ, Mallick SP, Penczek PA, Roseman AM, Sigworth FJ, Volkmann N, Potter CS., (2004). Automatic particle selection: results of a comparative study.. J Structural Biol. 145, pp.3-
Fourier transforms
What You Need To Know About A Fourier Transform
[edit | edit source]By David J DeRosier
Professor Emeritus,
Department of Biology & Rosenstiel Basic Medical Sciences Research Center,
Brandeis University
Introduction
[edit | edit source]Most students of molecular electron microscopy keep well away from learning about Fourier transforms. At schools whose aim is to train students in molecular electron microscopy, however, the gathered class must sit through a lecture or two on the Fourier transform. A mathematical lecture on the topic is usually more satisfying to the faculty than it is to students, who use the occasion to day dream or simply sleep having been up half the previous night at some bar. I guess it is not obvious why such a mathematical operation would be of interest to those who simply wants to know the molecular architecture of some cellular machine.
My aim is to tell you why you want a Fourier transform of your electron micrographs, what you can learn from a Fourier transform, how to think about a Fourier transform without having to waddle through the mathematics, and how to generate a Fourier transform when you want one. Since microscopy and image analysis are visual, I am presenting many of the lessons as pictures. I do not intend to prove the properties of various transforms but rather to show the results. There are lots of books on the theory.
Background on Fourier Transforms
[edit | edit source]Fourier Transforms are a tool used to analyze complicated data. They are, in essence, a mathematical function that transforms spatial or time based data into amplitudes and frequencies. This allows for the data to be analyzed at a glance, and more importantly makes it very easy to adjust features of the signal. Uses of the Fourier transforms include processing and analysis of sound, video, images, and other large, complex sources of data. The technology transition from analog technologies to digital has ushered in an increased use of this technique. Since the original algorithm for the transform is very long and computer resource intensive, a computer friendly version known as the Fast Fourier Transform (FFT) was independently invented by both J.W. Cooley and J.W. Tukey in 1965.
In transmission electron microscopy image analysis, fourier transforms are heavily utilized to remove the low resolution data from a collected image. Removing the low resolution data, which is the center of the image produced by a fast fourier transforms, is a way of increasing the contrast on images without losing much of the identifying information. It is often necessary to remove some of the high resolution data, which is found at the outside of the fast fourier transform, since the method of collection in a tunneling electron microscope often distorts the data. The need to do this step varies with the quality of the image recorded. For example, having a direct detector instead of film will greatly increase image quality and will allow more of the data transformed to be kept, which leads to a higher resolution 3D reconstruction.
Taking data from an electron microscopy image and transforming also allows for greater ease in 3D reconstruction. Fourier transforms are completely and easily reversible even after images have been processed, so frequently 3D reconstructions are generated with the fourier transforms of 2D images.
Fourier Transforms involve many of different types of math, but the most common element that is seen in fourier transform math is complex numbers. Complex numbers are usually seen in some form or another but a common form of this is . Now i stands for imaginary number. You might be asking yourself why involve such numbers or complex math altogether. Well its simple really, complex numbers make things go together really well. Rather than laying hundreds of equations and piling on thousands upon thousands of different mathematical proofs for you to see and understand, complex numbers shorten it and make it look a bit more elegant. Complex numbers is just one element that shows up in fourier equations and transforms.
Figure 1: Fourier Series formula, Purdue University.
Figure 1 listed above gives an apt description of what the formula for Fourier series looks like. Its expressed in sines and cosines, which are commonly found in frequencies. Fourier series and transforms allow any type of data to be converted into frequencies and amplitudes and it also allows us to delete certain frequencies away from the data. This allows us to clean up most of the data or image in our case. This is extremely helpful in electron microscopy as, many images taken from a EM are littered with aberrations or under focusing on the image. The fourier transform allows us to convert the images we see in to frequencies or amplitudes and allows us to modify or “delete” parts of the frequency away in order to clear up the image and correct any aberrations seen or found in the data. Then running an inverse of the fourier transform, turns the frequency back into an image with the corrections made.

Why you want a Fourier transform.
[edit | edit source]-
Caption1
-
Caption2
2D particle alignment
What is Alignment?
[edit | edit source]
Alignment is the process of rotating and shifting particles in order to position them in a similar direction. In order to obtain a proper averaged 2D image of the particle, the aligning process is used [1]. Most alignment methods require the particles to be centered. There are multiple approaches when it comes to 2D particle alignment. The main type of classification involves the accessibility to a reference image, dividing methodology into Reference-based alignment and Reference-free alignment
Reference Based Alignment
[edit | edit source]Reference Based Alignment involves a process of aligning every particle in its position and orientation with the reference image. In this approach the particles might be turned over and a mirror orientation needs to be applied. This is followed by an assessment of cross-correlation coefficient in order to establish the resemblance between a particle and a reference. There are many advantages to reference-based alignment. For instance, the technique is fast, effortless to perform, and since the reference is known it allows a visual verification of accurate orientation of the particle in interest. On the other hand, Reference Based Alignment also has drawbacks. The particle alignment can be biased. This is because of the assumption that the particle needs to look like the available reference image. Aligning the particle to faulty reference will result in a noisy data, which will look like the reference. This makes it difficult when evaluating for similarity. It is necessary to decipher if the resemblance is actual or if it is a product of the noise.
Reference-free Alignment
[edit | edit source]Reference-free Alignment is commonly used when there is no apparent assemblage in a data set and the reference images are not available. This particular technique looks for all the possible pairs in an entire data set that have the best comparative orientation. It is computed by applying the cross-correlation function. This method makes use of the random approximation algorithm of overall averages and iterations until convergence is reached. Reference-free alignment has certain advantages. The algorithm used in this method allows shift and rotational alignment of a series of images. It is also fast and requires only the radius of the particle. One of the main disadvantages of this method is the difficulty in aligning the particles that have multiple shapes, low signal to noise ratio or very small data sets. Technically, all reference free methods create a reference for processing data set.
Methods of Alignment
[edit | edit source]Classic Method of 2D Alignment
[edit | edit source]Classic Method of 2D Alignment is a reference based technique. It involves a shift in x and y coordinates, followed by an image rotation in plane, while making a comparison to the reference. Every rotation is tagged along by computation of cross-correlation between image and a reference. The highest cross-correlation value coincides with the best alignment.
Spider Method
[edit | edit source]Spider Method, also known as ring-based correlation, makes use of the rings. It takes different rings of pixels and cuts them up into little circles, lays them flat and stacks the rings in order to cross-correlate them. Instead of doing the x, y shift, the best correlation value is represented by the angle. This method is characterized by a lot of noise resulting in false positive.
Radon Transform
[edit | edit source]
Radon Transform is a useful in techniques like pattern recognition and image processing. It involves the cross sectional scans of the images. It takes the average to make a slice of the image after every rotation around its center. The stack of slices is the final radon transformation of the image. The next step in the process involves the angle approximation that can be calculated by the y shift of the radon transform, which corresponds to the angle of the reference image. This can also be done by using cross correlation in which the highest peak will give the best angle. The final step engages the rotation of the image to its reference. In addition to the averaging process, this method also calculates the error measures and resolution used in 2D alignment technique. Radon transform follows the 3D reconstruction process where translational and rotational alignments can be done in 2D and 3D [2].
Maximum Likelihood (ML)
[edit | edit source]Maximum Likelihood or ML is a reference free method, which involves finding the most probable model to fit in the obtained data. In this approach the probability of observations are measured. These are based on the original model and it is an alternative method to cross correlation technique that makes choice based on differences throughout the data set. ML method assesses the relative orientations of the particles, which are processed as concealed variables that are incorporated in likelihood computation. This ML refinement approach also considers the error model which in turn reduces the biased in the large data sets [3]. It is one of the slowest methods since it involves large data sets.
Classification
[edit | edit source]
Classification is an important step prior to the 3D reconstruction step. There is a strict requirement that individual images used for reconstruction should have. These are different views of the same biological specimen under study. It allows separating the different views of particles from a homogenous set of population. As in the case of a Gro-El molecule, classification applies as separating the top view and side view (Pascual-Montano 233-245).
Principal Component Analysis
[edit | edit source]Principal Component Analysis is used in image recognition and compression techniques. It is known as a mathematical method of grouping things together, followed by reducing their dimensionality, while preserving data that is closest to the original set. This attribute works well when there is a strong correlation between the recorded variable where it projects data along eigenvector. First it converts images into large matrix data. Next it analyzes the key features of different views and creates Eigen-images that have assigned Eigen values. The main functions of this method are to reduce redundancy, extract the general features, and make predictions of the images and data compression. This method is commonly used in the process of face recognition, where it finds the key features that separate the classes of images [4].
Self-Organizing Maps
[edit | edit source]Self-Organizing Maps are an excellent method of Data Mapping and Classifying when there is a large number of dataset and classes. This is useful, particularly if the boundaries between the classes are not clear. SOM is also called Kohonen Self Organizing Maps, because these were first discovered by Kohenen in 1997 [5]. SOM is a non-linear projection of high dimensional data in a low dimensional space. The structure of a neural network consists of 3 layers. These are the input layer, middle- hidden layer and the output layer. The SOM simulates the self-organizing process conducted by the human brain when input data is presented to it.
References
[edit | edit source]- ↑ "Methodology of 2D Particle Alignment." Wadsworth Center, [Online]. Accessed 16 Nov. 2013. Available from: http://spider.wadsworth.org/spider_doc/spider/docs/align.html
- ↑ Radermacher. M., (1997) Radon Transform techniques for Alignment and Three Dimensional Reconstruction from Random Projections. Scanning Microscopy Vol. 11, (171-177)
- ↑ Scheres.S.H.W., Valle.M., Nunez.R., Sorzano.C.O.S., Marabini.R., Herman.G.T., Carazao.J.M., (2005) Maximum-Likelihood Multi reference Refinement for Electron Microscopy Images. J. Mol. Biol. 348, (139–149)
- ↑ Kyungnam.K., “Face Recognition using Principal Component Analysis” University of Maryland, [Online]. Accessed 16 Nov. 2013. Available from: http://www.umiacs.umd.edu/~knkim/KG_VISA/PCA/FaceRecog_PCA_Kim.pdf
- ↑ Pascual-Montano A., Donate L.E., Valle M., Barcena M., Pascual-Marqui R.D., Carazo J.M., (2001) A Novel Neural Network Technique for Analysis and Classification of EM Single-Particle Images. Journal of Structural Biology 133,233-245
Initial model
Initial Modeling
[edit | edit source]Initial modeling is the process of using aligned and characterized electron microscopy images to produce a 3D reconstruction of the imaged particle. However, because each image is only a 2D projection of a 3D structure, it can often be quite difficult to determine the actual 3D structure of the original particle. To counteract this problem, researchers turn to the projection slice theorem: for any 3D object, the Fourier transform of each 2D projection it casts is a slice of the Fourier transform of the original 3D object. Thus, if enough 2D projections are obtained, the Fourier transform of the 3D object can be entirely reconstructed, and the original 3D object modeled in space by taking the inverse Fourier transform.
There are a number of common techniques used for initial modeling, some of which are described below.
Use of Generic Shapes
[edit | edit source]Information on generic structures of molecules can be useful for initial model formation. Using generic shapes like spheres, cubes etc. of molecules can help provide a starting point to obtain a detailed structure using a technique that has better resolution. It can often be easier to start with a generic shape and work towards a high-resolution model than to start from nothing and attempt to generate the model de novo. However, this method can introduce significant bias if the starting shape chosen is not an accurate representation of the actual structure of the molecule.
Use of Pre-existing Structures
[edit | edit source]Previously solved 3D structures are stored either in the Electron Microscopy Data Bank (EMDB) or the Protein Data Bank (PDB). 3D structures of biomolecules derived using electron microscopy are deposited in EMDB. EMDB’s for U.S, Japan, UK and Asia all act independently under their own protocols. Similarly, protein structures derived using X-ray crystallography, NMR, electron microscopy etc. are deposited at the Protein Data Bank (PDB). Approximately 90% structures submitted to PDB are derived using X-ray crystallography. The PDB contains the X, Y and Z coordinates for several protein structures.
New structures can be assessed against previous structures which helps validate the new structure in some capacity. When attempting to compile an initial model, a lower-resolution pre-existing structure is often used, so that any higher-resolution details are filled in by the obtained data in an attempt to avoid bias in the final structure. A structure can be taken from either the EMDBs or the PDB and blurred and then used to fill in the details of the new structure. For example in a research study while placing the protein and RNA structures into the 50S ribosomal subunit, the researchers used previously solved generic RNA-duplex and RNA tetraloops to position the density map of the 50S subunit [1]. Thus, lower resolution data can be combined with new high resolution data to facilitate better interpretation.
Common Lines Method
[edit | edit source]
The common lines method is a technique used to obtain an initial model by taking advantage of the points at which different projections of a structure intersect in Fourier space[2]. The creation of initial 3D models from 2D images acquired from single-particle electron microscopy indisputably relies on data obtained from different views of the molecule of interest. The central feature of the common lines method is the definitive relationship of the center section, 1D, in Fourier space of 2D image views in noise free space[2]
To obtain an initial 3D model reconstruction using the common lines method all particles are compared to one another in attempt to identify angular relationships. Since there are common lines for all particles the central section of the 2D Fourier transform can be determined by comparing the center section of the Fourier transform of all particles to one another. However, since the common lines method relies heavily on the identification of the shared 1D lines, the method is extremely hindered by noisy raw images. As a rule, the use of common lines on noisy raw images is error prone[3] . In addition to limitations related to signal-to-noise ratios, the common lines method cannot be used to identify the handedness of a molecule.
Random Conical Tilt
[edit | edit source]
Random Conical Tilt (RCT) requires that two images be taken of the same set of particles – one at a high tilt angle (usually around 60°), and the other at no tilt (0°) [4]. This results in two views of each particle within the sampling area that are physically related. The rotation angle of the molecule in the untilted image provides one angle for reconstruction, the angle of the specimen holder another, and the tilted images are used to construct the initial model [5]. These images can also be characterized and separated into distinct groups corresponding to different orientations of the imaged particles, resolving much of the uncertainty in samples of heterogeneous orientations [6].
However, RCT also has several limitations. The copper grid on which the sample sits limits the maximum angle to which the specimen can be tilted, and because the paired images are not orthogonal, there is a significant amount of missing data – the “missing cone” [4]. In addition, because two images of the same sample area are required, the sample is exposed to a much larger dose of radiation. The tilted image is usually taken first, and thus the images with the least radiation damage are used to reconstruct the image [5], but the alignment data must necessarily suffer, because it is taken from the second, untilted image [5]. This problem is more apparent when the samples are in vitreous ice (cryoEM) – both because the higher electron density of a negative stain serves to protect the sample particles from damage from the electron beam, and because the first dose of radiation can often melt the ice, causing the particles to drift between the first and second images. The vitreous ice of cryoEM samples is also often preserved by the use of a small carboy of liquid nitrogen on the end of the sample holder, and any movement of this carboy (such as that required to tilt the sample holder within the microscope) can cause the liquid nitrogen to boil, thus shaking the sample holder and reducing the resolution of the resulting images.
Orthogonal Tilt Reconstruction
[edit | edit source]Orthogonal Tilt Reconstruction (OTR) works in much the same way as RCT, with the exception that the paired images taken are at 90° angles to one another. Taking images at 0° and 90° is impossible, due to the physical dimensions of the sample holder (as mentioned previously), so OTR circumvents this problem by taking images at 45° and -45° [4]. The use of orthogonal images means that the entirety of the structure is sampled in Fourier space, thus eliminating the “missing cone” of RCT – this is its main advantage. However, it does still share the other limitations of RCT, as it still requires exposing a sample area to two doses of radiation and still requires physical movement of the sample holder.
OTR also has a unique limitation – it only works if the particles being imaged do not have a preferred orientation at 0° [4]; in other words, OTR works only for those particles that randomly assume all possible orientations on the sample area. For any particles which have a tendency to orient themselves in a certain way, OTR is not a viable reconstruction method.
Tomography
[edit | edit source]Tomography is again similar to both RCT and OTR, except that many more images are taken, at various angular orientations [7]. Again, the rotation angles are limited by the physical dimensions of the sample holder, meaning that some of the structure cannot be sampled – but unlike the “missing cone” of RCT, tomography results in a “missing wedge” [7]. Multiple rotation sets can be combined, however, to produce a complete initial model. Because many more images are required, though, the dosage of each image must be lowered dramatically to avoid radiation damage to the sample. Tomography thus requires a balancing act - maximizing the number of images that can be collected while still ensuring that each image is exposed to enough radiation to produce sufficient contrast for alignment [7].
Particle drift becomes a more significant problem in tomography, because there are so many more images in which the position of each particle must be determined. Thus, it is often beneficial to add markers of high electron-density to the particles, which will show up as constant dark spots in every image and can be used to track any motion of their associated particles in each image.
References
[edit | edit source]- ↑ Ban, N., Nissen, P., Hansen , J., Capel, M., Moore, B., & Steitz, T. (1999). Placement of protein and RNA structures into a 5 Å-resolution map of the 50s ribosomal subunit. Nature, (400), 841-847. doi: 10.1038/23641
- ↑ a b Joachim F. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State, 2nd edition. New York:Oxford University Press Inc, 2006
- ↑ Lyumkis D, Vinterbo S, Potter CS, Carragher B. Optimod - An automated approach for constructing and optimizing initial models for single-particle electron microscopy. J Struct Biol. 2013 Dec;184(3):417-26. doi: 10.1016/j.jsb.2013.10.009. Epub 2013 Oct 24. PubMed PMID: 24161732
- ↑ a b c d Yoshioka C, Pulokas J, Fellmann D, Potter CS, Milligan RA, Carragher B. Automation of random conical tilt and orthogonal tilt data collection using feature-based correlation. J Struct Biol [Internet]. Sep 2007 [cited 5 Dec 2013]; 159(3): 335-346. Available from: www.ncbi.nlm.nih.gov/pmc/articles/PMC2043090/
- ↑ a b c van Heel M, Orlova EV, Harauz G, Stark H, Dube P, Zemlin F, Schatz M. Angular reconstruction in three-dimensional electron microscopy: historical and theoretical aspects. Scanning Microscopy [Internet]. 1997 [cited 5 Dec 2013]; 11: 195-210. Available from: http://www.ecmjournal.org/journal/smi/pdf/smi97-15.pdf
- ↑ Chandramouli P, Hernandez-Lopez RA, Wang HW, Leschziner AE. Validation of the orthogonal tilt reconstruction method with a biological test sample. J Struct Biol [Internet]. 2011 [cited 5 Dec 2013]; 175(1): 85-96. Available from: http://dash.harvard.edu/bitstream/handle/1/8893763/Chandramouli_ValidationOrthogonal.pdf?sequence=1
- ↑ a b c Lučić V, Rigort A, Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. JCB [Internet]. 5 Aug 2013 [cited 5 Dec 2013]; 202(3): 407-19. Available from: http://jcb.rupress.org/content/202/3/407.full
3D Reconstruction
3D Reconstruction
[edit | edit source]
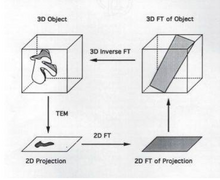



The goal of 3D reconstruction in transmission electron microscopy is to take a 2D projection and turn it into its 3D volume. This allows for the visualization of small structures within the sample. It is essential to reconstruct the 2D projection because the information gained from the projection itself is limited.
Projection
[edit | edit source]A projection occurs when a 3D image is taken and those points are then put onto a 2D plane. Each point in the 2D plane corresponds to a slice in the 3D volume and is based on Radon's theorem. It is possible to obtain a 3D volume, get the 2D projection, subsequently take the Fourier transform of the projection and use the Fourier transform of the projection to obtain a "slice" of the 3D volume. Thereafter, it is possible to take the inverse Fourier transform of the "slice" yielding a 3D volume.
Orientation of the Object
[edit | edit source]Before proceeding to reconstruction, it is necessary to know in which direction the particles are facing. The orientation of the particles can be determined by using their Euler Angles. Euler angles can be defined in two ways. According to the first definition, the orientation can be determined by using the latitude, longitude and rotation in place of the particle. The second definition takes into account the z and x axis. The initial position is determined using the Z axis. Next, the particle is rotated and the orientation is derived from the new X axis. The third and final calculation is based on the location of the new Z axis, after it has been rotated around its x axis.
What is a Backprojection?
[edit | edit source]A backprojection is basically the inverse of a Radon transform. It is the process where a 2D projection goes into a 3D model. It is the only method that can be used for this purpose. A backprojection reconstructs an image by taking each view and smearing it along the same path it was acquired (Fig. 1). The result from this is a blurry version of the correct image (Voss 2013)[1].
Resolution
[edit | edit source]Once a 3D reconstruction is obtained it is important to determine how good the reconstruction is. This is done by using the Fourier Shell Correlation. Essentially the primary set of particles is divided into an even subset and an odd subset in which a structure is calculated for each subset. Each set is then backprojected to obtain a volume for each subset. Since the goal is to determine the quality of the reconstruction the data is divided into rings within Fourier space with each ring representing different resolutions. The Correlation coefficient is then calculated between the structure for the even subset and the odd subset based on their frequency.

Footnotes
[edit | edit source]- ↑ Voss, Neil. 3D Reconstruction. Protein Structure 464. Roosevelt University, Schaumburg, IL. 25 Oct. 2013. Lecture.


