Structural Biochemistry/Protein Folding and Chaperones
Protein Folding
[edit | edit source]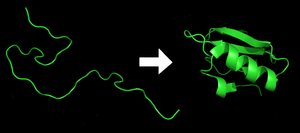
Protein folding is a highly complex process by which proteins are folded into their biochemically functional three-dimensional forms. The hydrophobic force is an important driving force behind protein folding. The polar side chains are usually directed towards and interact with water, while the hydrophobic core of the folded protein consists of non-polar side chains. Other forces that are favorable for protein folding are the formation of intramolecular hydrogen bonds and van der Waals forces.
Protein folding is entropically unfavorable because it minimizes the dispersal of energy and adds order to the system. However, the summation of the hydrophobic effect, hydrogen bonding, and van der Waals forces is greater in magnitude than the loss of entropy. Protein folding is therefore a spontaneous process because the sign of ΔG (Gibbs free energy) is negative.
For a reaction at constant temperature and pressure, the change ΔG in the Gibbs free energy is:
The sign of ΔG depends on the signs of the changes in enthalpy (ΔH) and entropy (ΔS), as well as on the absolute temperature (T, in kelvin). Notice that ΔG changes from positive to negative (or vice versa) where T = ΔH/ΔS.
When ΔG is negative, a process or chemical reaction proceeds spontaneously in the forward direction.
When ΔG is positive, the process proceeds spontaneously in reverse.
When ΔG is zero, the process is already in equilibrium, with no net change taking place over time.

Chaperones
[edit | edit source]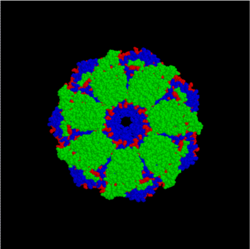
Intramolecular chaperones are essential for protein folding, but not required for protein function. Considerable evidence shows that chaperones play a critical role in protein folding both in vivo and in vitro. Unlike their molecular counterparts, intramolecular chaperones are encoded in the primary sequence of the protein as an N-terminal or C-terminal sequence extension and are usually termed propeptides or prosequences. Upon mediation of the protein folding, the intramolecular chaperones are removed either by auto-processing in the case of proteases or by an exogenous process in the case of non-protease proteins.
Intramolecular chaperones are classified into two groups on the basis of their roles in protein folding. The type I intramolecular chaperones mediate the folding of proteins into their respective tertiary structures and are mostly produced as the N-terminal sequence extension. The type II intramolecular chaperones mediate the formation of the quaternary or functional structure of proteins, and usually are located at the C-terminus of the protein.
Type I intramolecular chaperones
[edit | edit source]The discovery of the first intramolecular chaperone was based on the studies on subtilisin, an alkaline serine protease from bacillus subtilis. Often, the relation of intramolecular chaperones to the molecular mechanism of protein folding is studied by introducing amino acid substitution mutations in the propeptide region but not in the functional domain of the protein. It was shown that the addition of propeptides in trans allowed for the folding of the subtilisin at a higher efficiency and rate than when folded in cis. It was also shown that if the energy barrier of the transition state in subtilisin was reduced, it was allowed to fold without the intramolecular chaperone, but at a slower rate.
Distinct from the subtilisin protease, the NGF (nerve growth factor) propeptide forms a cysteine knot by virtue of three intramolecular disulfide bonds. Based on the hydrogen-deuterium exchange experiments and the spectroscopic studies, the propeptide acts as a competitive inhibitor for the receptor binding of the mature NGF dimer. It is likely that the quaternary structure may stabilize the tertiary structure.

It is suggested that the α-lytic protease folds through a nucleation mechanism, in which the propeptide folds first and acts as a scaffold that stabilizes the C-terminal domain of the mature protease. This allows for the structural arrangement of the two domains to pack into the native structure.
Sometimes the C-peptide has independent physiological functions. For example, the C-peptide of proinsulin both stimulates Na+, K+-ATPase and functions as an intramolecular chaperone for folding of insulin.
Type II intramolecular chaperones
[edit | edit source]Intramolecular chaperones that are involved in the folding of the quaternary structure of proteins are called type II intramolecular chaperones. The E. Coli K1-specific bacteriophages contain tail spikes that exist as homotrimers of endosialidases. These tail spikes are produced with a C-terminal domain (CTD) that is not part of the functional trimer. The fact that the CTD folds independently from the enzymatic domain and forms a hexamer suggests that the CTD is able to associate with each other to initiate the trimerization of endosialidases.

There exists both an N-terminal and a C-terminal propeptide in the fibril-forming collagen. The C-terminal propeptide prevents premature fibril formation, while the N-terminal propeptide is important in fibril associate of the collagen triple helix. The propeptide is proteolytically processed in the functional multimer.
Chaperones-protein folding in the cytoplasm of prokaryotic and eukaryotic cells
[edit | edit source]In vitro, many unfolded proteins attain the native state spontaneously, however within cells such folding efficiency is limited by the conditions. There are two factors expected of the cellular environment to improve propensity of polypeptides misfolding and aggregating. The Crowding Effect-with high concentration of macromolecules, and close proximity of emerging polypeptide chains developing from polyribosomes. A highly conserved set of proteins chaperones, prevented non-productive protein folding. In the cytoplasm, the Hsp 70 (heat-shockprotein of 70 kDa) and chaperones are the major factors to have efficient protein folding in normal conditions or adverse conditions (heat stress). Chaperonins interact with collapsed folding intermediates so efficient folding advance in the provided environment. For many proteins, both molecular chaperones are need for efficient folding.
Protein Misfolding
[edit | edit source]It is especially important to decipher the role intramolecular chaperones play in protein folding because human proteins are involved in diseases. These proteins are found to contain the sequence extensions that probably function as intramolecular chaperones. Mutations termed protein-memory mutations in the intramolecular chaperones can cause misfolding of the functional domain, which results in distortion of their function leading to human diseases.
The Future of Intramolecular Chaperone Research
[edit | edit source]The recent advantages of such technologies as single protein production (SPP) system, in-cell NMR, and cotranslational structural studies will serve as important techniques to further study protein structures and folding in the cell. Furthermore, the mechanisms of intramolecular chaperone-mediated protein folding in vivo can be investigated.
References
[edit | edit source]- C.B. Anfinsen, Principles that govern the folding of protein chains, Science 181 (1973), pp. 223–230.
- H. Ikemura, H. Takagi and M. Inouye, Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli, J Biol Chem 262 (1987), pp. 7859–7864.
- J.L. Silen, D. Frank, A. Fujishige, R. Bone and D.A. Agard, Analysis of prepro-alpha-lytic protease expression in Escherichia coli reveals that the pro region is required for activity, J Bacteriol 171 (1989), pp. 1320–1325.
- Dean J.Naylor and F.-Ulrich Hartl1, Department of Cellular Biochemistry, Max-Planck-Institut für Biochemie, Am Klopferspitz 18A, Martinsried bei München D-82152, Germany
