Proteomics/Post-translational Modification/Carboxyl Group Modifications
Chapter written by: Rhea Sanchez and Ashlee Benjamin
Contact ris6859@rit.edu or amb4541@rit.edu for contributions
This Section:
Carboxyl Group Modifications
[edit | edit source]Nearly all polypeptides undergo modifications after being translated from mRNA. Many types of these modifications occur at the ends of the polypeptide. The C-terminus of a protein or polypeptide is the end of the amino acid chain terminated by a free carboxyl group (-COOH)[1].
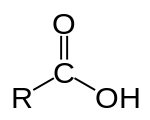
There are several ways in which the C-terminus of the polypeptide can be modified post-translationally. The most common post-translational modification is the addition of a lipid anchor to the C-terminus. This modification allows the protein to be anchored within the membrane without having a transmembrane domain [2].
Types of C-terminal Modification
[edit | edit source]There are several types of C-terminal modification including: amidation, prenylation, glypiation, ubiquitination, sumoylation, and methyl/ethyl-esterification.

Amidation
[edit | edit source]C-Terminal amidation is a common posttranslational modification in peptide hormones. Amidation is the addition of an amide group to the end of the polypeptide chain [3].
The amide group for C-terminal amidation is contributed by a Glycine residue. This Glycine is present in a precursor C-terminal sequence that resembles XGXX, where X represents any amino acid. When digesting the polypeptides with proteases like CpY (carboxypeptidase Y), CpA and CpB, CpY releases the C-terminal amides, but CpA and CpB do not [4]. Amidation neutralizes negative charges on the C-terminus of the polypeptide. Peptidylglycine α-hydroxylating monooxygenase and peptidyl α-hydroxyglycine α-amidating lyase are two enzymes associated with amidation, and both of which are encoded by the same gene: Peptidylglycine α-amidating monooxygenase [3].
Peptidylglycine α-amidating monooxygenase, or PAM, precursor encodes a multifunctional protein that can be divided into three parts. First, the NH2-terminal third of the PAM precursor contains the enzyme peptidylglycine α-hydroxylating monooxygenase, or PHM. Secondly, the inner third of the PAM precursor contains the enzyme peptidyl α-hydroxyglycine α-amidating lyase, or PAL. Finally, the COOH-terminal third of the PAM precursor encodes a transmembrane domain and a hydrophilic domain that may form a cytoplasmic tail [3].
Both activities successively catalyze the production of α-amidated peptides from their glycine-extended precursors in a two-step process.
Prenylation
[edit | edit source]Prenylation (also isoprenylation or lipidation) is the post-translational addition of hydrophobic molecules, called prenyl groups, to a protein. The process of prenylation is catalyzed by three enzymes: Farnesyltransferase and Geranylgeranyltransferase I and Geranylgeranyltransferase II [5].
Farnesyltransferase and Geranylgeranyltransferase I are structurally and functionally similar proteins. In terms of structure, both enzymes consist of two subunits: the α-subunit and the β-subunit. Both enzymes recognize the CAAX (C represents Cysteine; A represents an aliphatic amino acid; X represents any amino acid) box at the C-terminus of the target protein [5]. Depending on the enzyme, either a farnesyl-isoprenoid or geranylgeranyl-isoprenoid membrane anchor is added to the cysteine residue of the CAAX box near the C-terminus.
Geranylgeranyl transferase II, or Rab geranylgeranyltransferase, catalyzes a reaction in which geranylgeranyl groups are added to cysteine residues located near and/or at the C-terminal end of Rab proteins. However, due to the hypervariability of the C-termini of Rab proteins, Rab proteins do not have a consensus sequence, such as the CAAX box, which the Rab geranylgeranyl transferase can recognize. Thus, Rab proteins recruit the help of Rab escort proteins, or REP, which can then be recognized by the Rab geranylgeranyltransferase. After prenylation, the Rab proteins become insoluble due to the action of lipid anchors. REP is partially responsible for binding the geranylgeranyl groups and rendering them soluble. REP also transports the Rab protein to the appropriate membrane [5].
Glypiation
[edit | edit source]The C-terminus of the peptide consists of hydrophobic amino acids. The hydrophobic end of the peptide undergoes a proteolytic cleavage, and a glycosylphosphatidylinositol, or GPI anchor, is added to the C-terminus. This process is known as glypiation [6]. Enzymes called phospholipases then release the GPI-linked proteins. Phospholipase C is an enzyme known to hydrolyze the phosphodiester bond of the phosphatidylinositol found in GPI-anchored proteins, resulting in the release of the mature protein. GPI-anchors play a critical role in receptor-mediated signal transduction pathway. Thus, it is to no surprise that defects in the glypiation process or in phospholipase-catalyzed reaction can have dire effects on the organism. One condition associated with defective GPI anchors is the disease known as paroxysmal nocturnal hemoglobinuria [7]
Ubiquitination
[edit | edit source]
Ubiquitination, or ubiquitylation, is the process of covalently attaching one or more ubiquitin monomers via an isopeptide bond. Ubiquitination is most notably associated with targeting substrates for degradation by the 26S proteosome [8]. However, it has also been linked to post-translational protein localization, activity, conformation, and protein-protein interactions.
The precise role ubquitination plays in the fate of a protein is believed to depend on the length of the ubiquitin chain appended to the protein. Monoubiquitination is the attachment of a single ubiquitin monomer to a protein, a non-proteolytic signal involved in endocytosis, virus budding, endosomal sorting, DNA repair, histone regulation and export from the nucleus [9]. Seven lysine residues in ubiquitin affect its ability to attach other ubiquitin molecules. This process of attachment is known as polyubiquitination [9]. The best-studied examples are chains of four polyubiquitins linked through the Lysine residue at postion 48. These ubiquitins target proteins for degradation [8].
Deubiquitinating enzymes, or DUBs, can, however, can undo the modification by cleaving off the ubiquitin protein(s). Several disorders have been linked to ubiquitination including Angelman syndrome, Von Hippel-Lindau syndrome, Liddle's Syndrome, and Fanconi anemia.
Sumoylation
[edit | edit source]Small Ubiquitin-related Modifier proteins, or SUMO proteins, are small proteins that covalently attach to proteins in a process known as sumoylation. As its name suggests, SUMO proteins and ubiquitin are similar proteins sharing many characteristics including the sequence of enzymatic reactions directing either process. However, unlike ubiquitin, SUMO proteins do not act as proteolytic signals. SUMOylation has been associated with various cellular functions and processes including cell cycle regulation, DNA repair, chromosomal maintenance, modifying cytoplasmic signal transduction, nuclear import and subnuclear compartmentalization, DNA repair, transcription regulation,and stress response [10][11].
Methyl-Esterification
[edit | edit source]Methyl esterification involves the conversion of carboxylic acids to methyl esters [12]. Methyl esterification has been associated with the membrane localization of proteins including small G proteins [13]. Carboxyl methyl esters have also been linked to rendering peptides resistant to exopeptidases [14].
Function
[edit | edit source]Post-translational modification of the Carboxyl terminus of a polypeptide has been linked to many biological functions and processes as listed below:
- Peptide activation
- Membrane integration
- Localization
- Activity
- Structure
- Protein-protein interactions
- Proteolytic signalling
- Endocytosis
- Endosomal sorting
- Histone regulation
- DNA repair
- Virus budding
- Nuclear export
- Cell cycle regulation
- Chromosomal maintenance
- Modifying cytoplasmic signal transduction
- Nuclear import and subnuclear compartmentalization
- Transcription regulation
- Stress response
References
[edit | edit source]- ↑ Wikipedia. “C-terminus.” Created: 28 November 2007. Accessed: 10 March 2008. Available at: <http://en.wikipedia.org/wiki/C-terminus>
- ↑ Wikipedia. “Post-translational Modification.” Created: 16 February 2008. Accessed: 10 March 2008. Available at: <http://en.wikipedia.org/wiki/Posttranslational_modification>
- ↑ a b c Driscoll WJ, Mueller SA, Eipper BA, and Mueller GP. “Differential Regulation of Peptide-Amidation by Dexamethasone and Disulfiram.” Molecular Pharmacology 1999; 55:1067-1076. [Online Source]
- ↑ Harris R. “Identification of Post-translational Modifications of Proteins.” Created: 11 September 1995. Accessed: 10 March 2008. [Online Source]
- ↑ a b c Wikipedia. “Prenylation.” Created: 25 March 2008. Accessed: 28 March 2008. Available at: <http://en.wikipedia.org/wiki/Prenylation>
- ↑ Coussen F, Ayon A, Le Goff A, Leroy J, Massoulié J, and Bo S. "Addition of a Glycophosphatidylinositol to Acetylcholinesterase." J. Biol. Chem. 2001; 276:27881-27892. [Online Source]
- ↑ Wikipedia. “Glycophosphatidylinositol.” Created: 7 January 2008. Accessed: 28 March 2008. Available at: <http://en.wikipedia.org/wiki/Glycophosphatidylinositol>
- ↑ a b Pickart CM. "Targeting of substrates to the 26S proteasome." FASEB Journal. 1997; 11: 1055-1066. [Online Source]
- ↑ a b Woelk T, Sigismund S, Penengo L and Polo S. "The ubiquitination code: a signalling problem." Cell Division. 2007; 2:pp 11. [Online Source]
- ↑ Saracco SA, Miller MJ, Kurepa J, and Vierstra RD. "Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential." Plant Physiol. 2007; 145:119–134. [Online Source]
- ↑ Kroetz MB. "SUMO: a ubiquitin-like protein modifier." Yale J Biol Med. 2005; 78:197-201. [Online Source]
- ↑ Ma M, Kutz-Naber KK, and Li L. "Methyl Esterification Assisted MALDI FTMS Characterization of the Orcokinin Neuropeptide Family." Anal. Chem. 2007; 79: 673-681. [Online Source]
- McFadden PN and Clarke S. "Chemical Conversion of Aspartyl Peptides to Isoaspartyl Peptide."J Biol Chem. 1986; 261:11503-11511. [Online Source]
- ↑ Fujiyama A, Tsunasawa S, Tamanoi F, and Sakiyama F. "S-farnesylation and methyl esterification of C-terminal domain of yeast RAS2 protein prior to fatty acid acylation." J Biol Chem. 1991; 266:17926-17931. [Online Source]
- ↑ McFadden PN and Clarke S. "Chemical Conversion of Aspartyl Peptides to Isoaspartyl Peptide."J Biol Chem. 1986; 261:11503-11511. [Online Source]


