Methods and Concepts in the Life Sciences/Vectors
Vectors
[edit | edit source]In molecular cloning, a vector is a DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids.
The vector itself is generally a DNA sequence that consists of an insert (transgene) and a larger sequence that serves as the "backbone" of the vector. The purpose of a vector which transfers genetic information to another cell is typically to isolate, multiply, or express the insert in the target cell.
Cloning vectors are used to amplify DNA fragments. The basic features of a cloning vector are an origin of replication, a selectable marker (usually an antibiotic resistance) and a multiple cloning site (MCS, or polylinker). A multiple cloning site is a short region containing several commonly used restriction sites, allowing the easy insertion of DNA fragments at this location. On top of that, some cloning vectors carry other features such as a reporter gene which facilitates the screening for successful clones. The most prominent example is the lacZα gene, which can be used for α-complementation in blue-white screening.
Expression vectors are used to produce proteins. For this reason, they must carry some additional features, most importantly a promoter, a ribosome binding site and a terminator.
Plasmids
[edit | edit source]
Plasmids are extrachromosmomal DNA molecules which are found in essentially all types of bacteria. Most commonly, they are circular molecules of double-stranded DNA, although linear plasmids exist as well. Their size can vary from a few thousand to hundreds of thousands of base pairs. In nature, plasmids carry genes that may benefit survival of the organism (e.g. an antibiotic resistance), however, they are usually not essential for bacterial growth under normal conditions. Frequently, plasmids can be transmitted from one bacterium to another (even of another species) via horizontal gene transfer. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms.
Replication
[edit | edit source]Plasmids are replicons, which means they can replicate autonomously. However, they mostly rely on the DNA replication machinery of the host cell to do so. The region of the plasmid in which the replication starts is called the origin of replication (abbreviated ori or rep, sometimes also oriV for vegetative origin to distinguish it from the oriT which is necessary for conjugation). The origin of replication determines the replication mechanism, of which there are two general variants. Proteins which are needed for replication are usually encoded in close proximity to the ori. Apart from this, the ori is also the decisive element for other properties of the plasmid, such as its copy number, host range and incompatibility group.
Theta replication
[edit | edit source]The most common replication mechanism is called theta replication. It starts with the separation of the two strands at the ori region, which creates a structure that resembles the Greek letter theta (θ). The replication begins at an RNA primer and then proceeds either in one or both directions around the plasmid. The DNA molecules separate once the replication fork reaches the ori again (unidirectional) or the two replication forks meet (bidirectional).
The plasmid ColE1 and many cloning vectors whose oris have been derived from this plasmid show this mechanism.
Rolling-circle replication
[edit | edit source]Rolling-circle replication is another widespread replication mechanism. In this case, replication consists of two stages: At first, DNA is replicated to form both a double-stranded and a single-stranded circular DNA. Afterwards, the complementary strand is added to the single-stranded DNA, resulting in two double-stranded plasmids.
Rolling-circle replication is initiated by the Rep protein, which is encoded by the plasmid. It binds to the double-strand origin (DSO) of the plasmid, which might allow the DNA to form a cruciform structure. The protein then nicks one of the strands and remains covalently bound to the 5' phosphate end of the nicked strand. The free 3' hydroxyl end is released to serve as a primer for DNA synthesis by DNA polymerase III. Using the unnicked strand as a template, replication proceeds around the plasmid, displacing the nicked strand as single-stranded DNA. Once this single-stranded circle is complete, the 5' end which is attached to Rep and the displaced 3' end are ligated in a phosphotransferase reaction. The double-stranded circle is closed as well when the DNA Polymerase returns to the DSO. This probably involves creating a second nick and the host DNA ligase.
The replication of the displaced single strand starts at the single-strand origin (SSO), which is only synthesized when the displacement is almost complete. At the SSO, the RNA polymerase makes a primer for DNA polymerase III, which synthesizes the complementary strand. Once it is finished, DNA polymerase I replaces the RNA primer and DNA ligase joins the ends.

Copy number
[edit | edit source]The number of a particular plasmid per cell is a characteristic determined mostly by the origin of replication. It can vary from only one to several hundred copies of the same plasmid. Natural plasmids tend to have a low copy number, but it can be increased drastically by mutations and the elimination of control mechanisms. Controlling the copy number is very important for plasmids. On the one hand, they must keep up with the replication of the host cell. On the other hand, they should not become too big a burden for their host. Plasmids with high copy numbers only need a mechanism for the latter case, which means that replication should be inhibited once a certain copy number is reached. Such plasmids are called relaxed plasmids. Stringent plasmids, on the contrary, have a low copy number and therefore need a tighter control mechanism.
Control of replication at ColE1 origin
[edit | edit source]The copy number of plasmids with the ori of ColE1 is regulated by two RNA transcripts, RNA II and RNA I. RNA II is needed for the initiation of replication, whereas RNA I acts as its repressor.
The nascent RNA II, whose transcription is initiated 555 bp upstream of the replication origin, forms a hybrid with the template DNA near the ori. RNase H cleaves the RNA strand and exposes a 3' hydroxyl group. The processed RNA II can then serve as a primer for DNA synthesis by DNA Polymerase I.
The primer formation is inhibited by RNA I, a shorter transcript which is complementary to the 5` end of RNA II. When RNA I is present, it forms a hybrid with RNA II. This alters the folding of RNA II so that the DNA-RNA hybrid is not stabilized and cleavage does not occur. The rate of degradation of RNA I is therefore a major factor in control of plasmid replication. This rate of degradation is aided by the pcnB (plasmid copy number B) gene product, which polyadenylates the 3' end of RNAI targeting it for degradation by PNPase.
The binding of RNA I to RNA II is enhanced by the rop protein. Deletion of the corresponding gene therefore increases the plasmid copy number.

Host range
[edit | edit source]The types of bacteria a plasmid can replicate in is called its host range. It is usually determined by the origin of replication. Plasmids with the ColE1 ori have a narrow host range, they can merely replicate in E. coli and some closely related species. In contrast to this, broad-host-range plasmids can replicate in many bacteria which are only distantly related. These plasmids encode all proteins that are necessary for the initiation of replication so that they are independent of the host cell in this regard. Furthermore, their promotors and ribosome binding sites are recognized in a large number of bacteria.
Determining the host range of a plasmid can be challenging, especially when it comes to transformation and selection. For this reason, the actual host range of most plasmids is unknown.
Incompatibility
[edit | edit source]Two plasmids are incompatible when they cannot coexist stably in the same cell. This applies to plasmids with the same origin of replication, since they compete for replication factors. If one is replicated faster (for example because it is smaller) or has a different growth advantage, it will outgrow the other over a number of generations. Unequal distribution of the two plasmids during cell division can lead to curing of one of them as well.
By analyzing if the probability of curing increases when two particular plasmids are present in the same cell, one can assign plasmids to incompatibility (Inc) groups. While members of the same Inc group cannot coexist over a long time, plasmids from different groups can coexist stably and are not cured more frequently than in the absence of the other plasmid.
Partitioning and plasmid maintenance
[edit | edit source]Plasmids are a metabolic burden for their host cells. Consequently, plasmid-free cells usually have a growth advantage and would supersede plasmid-containing cells in the long term. To avoid this, different mechanisms have evolved which increase the segregational stability of plasmids, i.e. the likelihood that a plasmid is passed on from one generation of cells to another. This is not to be confused with structural stability, which refers to the preservation of the nucleotide sequence.
Random partitioning
[edit | edit source]In the simplest case, plasmids are passed on randomly to the daughter cells. The higher the copy number, the likelier it is that both cells inherit a plasmid. When the copy number is low however, it is likely that plasmid-free cells will develop and outgrow the other cells, unless other mechanisms prevent this.
Active partitioning
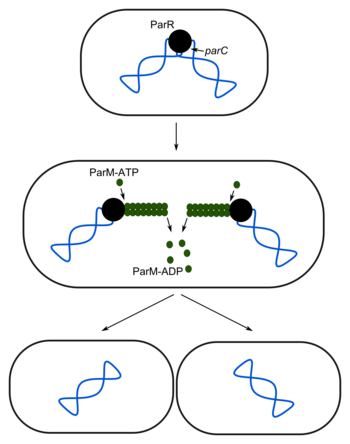
[edit | edit source]An effective way of ensuring that both cells receive a copy of the plasmid during cell division is a partitioning system. Such systems can be found on plasmids which are present in only one or very few copies per cell. Their replication is controlled in a similar manner as that of the chromosome, which means that the plasmid replicates every time the chromosomal replication is initiated. The best understood example is the Par system of the R1 plasmid of E. coli.
This system consists of the proteins ParR and ParM and the parC site on the plasmid. After replication, ParR interacts with the parC sites of the plasmids to form a partitioning complex. ParM-ATP binds to this complex and forms a filament, which pushes the plasmids towards the poles. ATP is hydrolyzed and ParM-ADP depolymerizes, leaving the plasmids at opposite poles.
Selective advantages
[edit | edit source]Many plasmids carry at least one gene which provides their host with a selective advantage. Most often, this is an antibiotic resistance. Another possibility is that the plasmid contains an important gene which is mutated or deleted in the host´s chromosome. Under the corresponding selection pressure, cells will retain these plasmids because cured cells grow slowly or die.
Addiction systems
[edit | edit source]Some plasmids include a system which kills the cell in case it does not inherit a plasmid during cell division. These toxin-antitoxin systems consist of two or more closely linked genes that encode both a 'poison' and a corresponding 'antidote'. If the plasmid is absent after cell division, the unstable anti-toxin is degraded and the stable toxic protein kills the new cell. This is known as post-segregational killing (PSK).
Some toxin-antitoxin systems rely on the base-pairing of complementary antitoxin RNA with the toxin's mRNA. Translation of the mRNA is then inhibited either by degradation via RNase III or by occluding the Shine-Dalgarno sequence or ribosome binding site. Often the toxin and antitoxin are encoded on opposite strands of DNA.
In other systems a labile protein antitoxin tightly binds and inhibits the activity of a stable toxin. An example is the CcdA/CcdB system which can be found on the F plasmid of Escherichia coli. CcdB targets the DNA gyrase and inhibits partitioning of the chromosomal DNA. As long as the cell contains the F plasmid, CcdB is bound and neutralized by CcdA. Upon loss of the plasmid, CcdA decays and the activity of CcdB kills the cell.
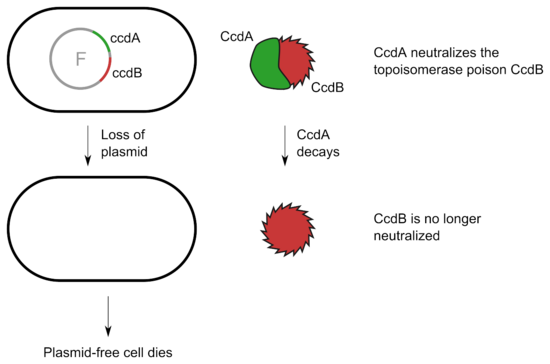
In biotechnology, toxin-antitoxin systems can be used to maintain plasmids in a bacterial culture. Furthermore, they can be used in molecular cloning to positively select for cells that have taken up a plasmid containing an insert. An example of this application comes from the ccdB gene, which has been incorporated into plasmid vectors. The gene of interest is then targeted to recombine into the ccdB locus, inactivating the transcription of the toxic protein. Thus, cells containing the plasmid but not the insert perish due to the toxic effects of the CcdB protein, and only those that incorporate the insert survive.
Resolution of multimeric plasmids
[edit | edit source]Plasmids can form dimers or higher multimers, which lowers the effective copy number and thus increases the risk that a cell does not receive a plasmid during cell division. Dimers are probably the result of recombination between two monomers. Subsequent recombination can then form higher multimers. To avoid the accumulation of multimers, many plasmids have site-specific recombination systems which can resolve multimers. These systems consist of proteins, which may be encoded by the host or the plasmid, and specific sites on the plasmid. In a dimer or multimer, the recombination site occurs more than once. The system then promotes recombination between these sites, which revolves multimers into monomeric plasmids.
A well-known example is the cer-XerCD system used by ColE1. cer is the recombination site on the plasmid, while XerC and XerD are part of a site-specific recombination system in E. coli. The cer site is only recognized if two other host proteins, PepA and ArgR, are bound close to it. If this is the case, the XerCD recombinase can promote recombination between two cer sites.
Conformations
[edit | edit source]Plasmid DNA may appear in one of five conformations, which (for a given size) run at different speeds in a gel during electrophoresis. The conformations are listed below in order of electrophoretic mobility (speed for a given applied voltage) from slowest to fastest:
- Nicked open-circular DNA has one strand cut.
- Relaxed circular DNA is fully intact with both strands uncut, but has been enzymatically relaxed (supercoils removed).
- Linear DNA has free ends, either because both strands have been cut or because the DNA was linear in vivo.
- Supercoiled (or covalently closed-circular) DNA is fully intact with both strands uncut, and with an integral twist, resulting in a compact form.
- Supercoiled denatured DNA is like supercoiled DNA, but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation.
Plasmids in molecular biology
[edit | edit source]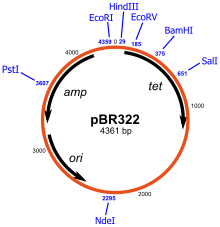
Plasmids are the most common vectors in genetic engineering. They serve as important tools in genetics and biotechnology labs, where they are commonly used to multiply or express particular genes. Many plasmids are commercially available for such uses.
High-copy plasmids are suited as cloning vectors. Expression vectors on the other hand can be low-copy plasmids, since the focus is on the production of protein instead of DNA.
Examples of common cloning vectors are pBR322 and pUC19. pBR322 was created in 1977 and was one of the first widely used E. coli cloning vectors. It was named after the Mexican postdoctoral researchers who constructed it. The p stands for "plasmid", and BR for "Bolivar" and "Rodriguez". pBR322 is 4361 base pairs in length and contains the ori of pMB1, a close relative of ColE1. It encodes two proteins which make E. coli resistant against ampicillin and tetracyclin and has unique restriction sites for more than forty restriction enzymes.

pUC19 is one of the most widely used plasmid vectors. It contains an ampicillin resistance gene (ampR) and the lacZα fragment for blue-white screening. The ori site is derived from pMB1. pUC is small but has a high copy number, which is a result of the lack of the rop gene and a single point mutation in the ori of pMB1. pUC18, another popular cloning vector, differs from pUC19 only in the orientation of the MCS.
Phagemids
[edit | edit source]A phagemid or phasmid is a plasmid that contains an f1 origin of replication from an f1 phage. Many commonly used plasmids contain an f1 ori and are thus phagemids. The f1 ori enables single stranded replication and packaging into phage particles. However, phagemids also contain an origin of replication for double stranded replication, which means that they can be replicated as a plasmid.
Similarly to a plasmid, a phagemid can be used to clone DNA fragments and be introduced into a bacterial host. However, infection of a bacterial host containing a phagemid with a 'helper' phage, for example VCSM13 or M13K07, provides the necessary viral components to enable single stranded DNA replication and packaging of the phagemid DNA into phage particles. Filamentous phages retard bacterial growth but, contrasting with the lambda phage and the T7 phage, are not generally lytic. Helper phages are usually engineered to package less efficiently (via a defective phage ori) than the phagemid so that the resultant phage particles contain predominantly phagemid DNA. F1 filamentous phage infection requires the presence of a pilus so only bacterial hosts containing the F-plasmid or its derivatives can be used to generate phage particles.
Prior to the development of cycle sequencing, phagemids were used to generate single stranded DNA templates for sequencing purposes. Today phagemids are still useful for generating templates for site-directed mutagenesis. Detailed characterisation of the filamentous phage life cycle and structural features lead to the development of the phage display technology, in which a range of peptides and proteins can be expressed as fusions to phage coat proteins and displayed on the viral surface. The displayed peptides and polypeptides are associated with the corresponding coding DNA within the phage particle and so this technique lends itself to the study of protein-protein interactions and other ligand/receptor combinations.
Cosmids
[edit | edit source]A cosmid is a type of hybrid plasmid that contains a lambda phage cos sequence (cos sites + plasmid = cosmids). Cosmids can replicate as plasmids if they have a suitable origin of replication: for example SV40 ori in mammalian cells, ColE1 ori for double-stranded DNA replication or f1 ori for single-stranded DNA replication in prokaryotes. Unlike plasmids, they can also be packaged in phage capsids, which allows the foreign genes to be transferred into or between cells by transduction.
Plasmids become unstable after a certain amount of DNA has been inserted into them, because their increased size is more conducive to recombination. To circumvent this, phage transduction is used instead. This is made possible by the cohesive ends, also known as cos sites. In this way, they are similar to using the lambda phage as a vector, with the difference that all lambda genes have been deleted except for the cos sequence. Cosmids can contain 37 to 52 (normally 45) kb of DNA, limits based on the normal bacteriophage packaging size. Cosmids can be used to build genomic libraries.
Bacterial artificial chromosomes
[edit | edit source]A bacterial artificial chromosome (BAC) is based on a functional fertility plasmid (or F-plasmid), used for transforming and cloning in bacteria. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150-350 kb. A similar cloning vector called a PAC has also been produced from the bacterial P1-plasmid.
BACs were frequently used in genome projects such as the Human Genome Project. In this procedure, a short piece of the organism's DNA is amplified as an insert in BACs, and then sequenced. Finally, the sequenced parts are rearranged in silico, resulting in the genomic sequence of the organism. BACs were replaced with faster and less laborious sequencing methods like whole genome shotgun sequencing and more recently next-gen sequencing.
Yeast artificial chromosomes
[edit | edit source]Yeast artificial chromosomes (YACs) are genetically engineered chromosomes derived from the DNA of Saccharomyces cerevisiae, which is then ligated into a bacterial plasmid. By inserting large fragments of DNA, from 100–1000 kb, the inserted sequences can be cloned and physically mapped using chromosome walking. This is the process that was initially used for the Human Genome Project, however due to stability issues, YACs were abandoned for the use of bacterial artificial chromosomes. Beginning with the initial research of the Rankin et al., Strul et al., and Hsaio et al., the inherently fragile chromosome was stabilized by discovering the necessary autonomously replicating sequence (ARS); a refined YAC utilizing this data was described in 1983 by Murray et al. The primary components of a YAC are the ARS, centromere, and telomeres from S. cerevisiae. Additionally, selectable marker genes, such as antibiotic resistance and a visible marker are utilized to select transformed yeast cells. Without these sequences, the chromosome will not be stable during extracellular replication, and would not be distinguishable from colonies without the vector.
Yeast expression vectors, such as YACs, YIps (yeast integrating plasmids), and YEps (yeast episomal plasmids), have an advantage over bacterial artificial chromosomes in that they can be used to express eukaryotic proteins that require posttranslational modification. By being able to inset large fragments of DNA, YACs can be utilized to clone and assemble the entire genomes of an organism. With the insertion of a YAC into yeast cells, they can be propagated as linear artificial chromosomes, cloning the inserted regions of DNA in the process.
Common features of bacterial vectors
[edit | edit source]Antibiotic resistance
[edit | edit source]Ampicillin
[edit | edit source]Belonging to the penicillin group of beta-lactam antibiotics, ampicillin is able to penetrate Gram-positive and some Gram-negative bacteria. It differs from penicillin G, or benzylpenicillin, only by the presence of an amino group. That amino group helps the drug penetrate the outer membrane of Gram-negative bacteria.
Ampicillin acts as an irreversible inhibitor of the enzyme transpeptidase, which is needed by bacteria to make their cell walls. It inhibits the third and final stage of bacterial cell wall synthesis in binary fission, which ultimately leads to cell lysis. Ampicillin is bacteriocidal.
Resistance to ampicillin is conferred by the amp or bla gene, which encodes a beta-lactamase. Beta-lactamases provide resistance to β-Lactam antibiotics by breaking the antibiotics' structure.

In E. coli, beta-lactamase is secreted into the periplasm and can be released into the culture medium, where it quickly degrades ampicillin. On plates, satellite colonies can form when the ampicillin concentration is too low.
Kanamycin
[edit | edit source]Kanamycin is an aminoglycoside bacteriocidal antibiotic, which interacts with the 30S subunit of prokaryotic ribosomes. It induces substantial amounts of mistranslation and indirectly inhibits translocation during protein synthesis.
Resistance to kanamycin is conferred by the npt II gene (also dubbed kan or neo) from the transposon Tn5, which encodes a neomycin phosphotransferase.
Streptomycin
[edit | edit source]Streptomycin was the first aminoglycoside antibiotic to be discovered, and it was the first cure for tuberculosis. Streptomycin inhibits the protein synthesis of both Gram-positive and Gram-negative bacteria. It binds to the small 16S rRNA of the 30S subunit of the bacterial ribosome, interfering with the binding of formyl-methionyl-tRNA to the 30S subunit. This leads to codon misreading, eventual inhibition of protein synthesis and ultimately death of microbial cells.
Chloramphenicol
[edit | edit source]Chloramphenicol is a bacteriostatic antibiotic effective against a wide variety of Gram-positive and Gram-negative bacteria, including most anaerobic organisms. It prevents protein chain elongation by inhibiting the peptidyl transferase activity of the bacterial ribosome.
The cat gene confers resistance to chloramphenicol. This gene codes for an enzyme called chloramphenicol acetyltransferase, which inactivates chloramphenicol by covalently linking one or two acetyl groups, derived from acetyl-S-coenzyme A, to the hydroxyl groups on the chloramphenicol molecule. The acetylation prevents chloramphenicol from binding to the ribosome.
Tetracycline
[edit | edit source]Tetracycline is a bacteriostatic antibiotic which inhibits protein synthesis by blocking the attachment of charged aminoacyl-tRNA to the A site on the ribosome. Tetracycline binds to the 30S subunit of microbial ribosomes. Thus, it prevents the introduction of new amino acids to the nascent peptide chain. The action is usually inhibitory and reversible upon withdrawal of the drug. Mammalian cells are less vulnerable to the effect of tetracyclines, despite the fact that tetracycline binds to the small ribosomal subunit of both prokaryotes and eukaryotes (30S and 40S respectively). This is because bacteria actively pump tetracycline into their cytoplasm, whereas mammalian cells do not.
Several resistance genes are used in molecular biology, the most common being tetA, tetB and tetC, which encode an efflux system, i.e. a membrane-associated protein which actively exports tetracycline from the cell. These genes are regulated by a repressor protein (TetR). This feature has been exploited to control gene expression (Tet-On and Tet-Off).
lacZ
[edit | edit source]
β-galactosidase is a protein encoded by the lacZ gene of the lac operon, and it exists as a homotetramer in its active state. However, a mutant β-galactosidase derived from the M15 strain of E. coli has its N-terminal residues 11—41 deleted and this mutant, the ω-peptide, is unable to form a tetramer and is inactive. This mutant form may, however, return fully to its active tetrameric state in the presence of an N-terminal fragment of the protein, the α-peptide. The rescue of function of the mutant β-galactosidase by the α-peptide is called α-complementation.
In this method of screening, the host E. coli strain carries the lacZ deletion mutant (lacZΔM15} which contains the ω-peptide, while the plasmids used carry the lacZα sequence which encodes the first 59 residues of β-galactosidase (the α-peptide). Neither is functional by itself. However, when the two peptides are expressed together, as when a plasmid containing the lacZα sequence is transformed into lacZΔM15 cells, they form a functional β-galactosidase enzyme.
The blue-white screening method works by disrupting this α-complementation process. The plasmid carries an internal multiple cloning site within the lacZα sequence. The integration of an insert into the MCS disrupts the gene, thus no functional α-peptide can be produced. Consequently, in cells containing the plasmid with an insert, no functional β-galactosidase may be formed.
The presence of an active β-galactosidase can be detected by X-gal, a colorless analog of lactose that may be cleaved by β-galactosidase to form a bright blue insoluble pigment. This results in a characteristic blue colour in cells containing a functional β-galactosidase. Blue colonies therefore show that they may contain a vector with an uninterrupted lacZα (therefore no insert), while white colonies, where X-gal is not hydrolyzed, indicate the presence of an insert in lacZα which disrupts the formation of an active β-galactosidase.
Common features of mammalian vectors
[edit | edit source]Promoters
[edit | edit source]CMV
[edit | edit source]The immediate-early Cytomegalovirus virus promoter can be used for high-level expression in a wide variety of mammalian cell lines.
SV40
[edit | edit source]The SV40 (Simian Virus 40) promoter contains the SV40 enhancer promoter region and origin of replication for high-level expression and replication in cell lines expressing the large T antigen. It contains two 72 bp SV40 enhancer repeats.
EF-1α
[edit | edit source]The EF-1α promoter is a strong, constitutive non-viral promoter. The EF-1α gene encodes the Elongation Factor-1α that catalyzes the GTP-dependent binding of aminoacyl-tRNA to ribosomes. EF-1α is one of the most abundant proteins in eukaryotic cells and is expressed in almost all kinds of mammalian cells.
UbC
[edit | edit source]The UbC (Human Ubiquitin C) promoter provides high-level expression across a broad range of species and tissue types. In HEK293 cells, it typically has about 50% of the activity of CMV and EF-1α promoters.
PGK
[edit | edit source]The PGK (Murine Phosphoglycerate Kinase-1) promoter is a ubiquitous housekeeping promoter which promotes long-term persistent expression.
Kozak sequence
[edit | edit source]
The Kozak consensus sequence is a sequence which occurs on eukaryotic mRNA and plays a major role in the initiation of the translation process. It has the consensus (gcc)gccRccAUGG. A lower case letter denotes the most common base at a position where the base can nevertheless vary while upper case letters indicate highly conserved bases. The sequence in brackets is of uncertain significance. The nucleotides AUG are the initiation codon encoding a methionine amino acid at the N-terminus of the protein.
The ribosome requires this sequence, or a possible variation to initiate translation. Variations can affect the amount of protein which is synthesized from the mRNA.
The Kozak sequence is not to be confused with the ribosomal binding site (RBS).
Selectable markers
[edit | edit source]G418
[edit | edit source]G418 (Geneticin) is an aminoglycoside antibiotic which blocks polypeptide synthesis by inhibiting the elongation step in both prokaryotic and eukaryotic cells. Resistance to G418 is conferred by the neo gene from Tn5 encoding an aminoglycoside 3'-phosphotransferase, APT 3' II. G418 is commonly used in laboratory research to select genetically engineered cells (typically using the KanMX selectable marker). In general, for bacteria and algae concentrations of 5 mg/L or less are used, for mammalian cells concentrations of approximately 400 mg/L are used for selection and 200 mg/L for maintenance. However, optimal concentration for resistant clones selection in mammalian cells depends on the cell line used as well as on the plasmid carrying the resistance gene, therefore antibiotic titration should be done to find the best condition for every experimental system.
Blasticidin S
[edit | edit source]Blasticidin S prevents the growth of both eukaryotic and prokaryotic cells. It works by inhibiting the termination step of translation and (to a lesser extent) the peptide bond formation by the ribosome. The most common resistance genes are bsr and bsd, which both encode a deaminase.
Hygromycin B
[edit | edit source]Hygromycin B is an antibiotic produced by the bacterium Streptomyces hygroscopicus. It is an aminoglycoside that kills bacteria, fungi and higher eukaryotic cells by inhibiting protein synthesis. It stabilizes the tRNA-ribosomal acceptor site, thereby inhibiting translocation. The resistance gene (hyg or hph) is a kinase that inactivates hygromycin B through phosphorylation.
Puromycin
[edit | edit source]Puromycin is an aminonucleoside antibiotic, derived from the Streptomyces alboniger bacterium, that causes premature chain termination during translation taking place in the ribosome. Part of the molecule resembles the 3' end of the aminoacylated tRNA. It enters the A site and transfers to the growing chain, causing the formation of a puromycylated nascent chain and premature chain release.
Resistance to puromycin is conferred by the pac gene encoding a puromycin N-acetyl-transferase (PAC) that was found in a Streptomyces producer strain. Puromycin is soluble in water (50 mg/ml) as colorless solution at 10 mg/ml. Puromycin is stable for one year as solution when stored at -20 °C. The recommended dose as a selection agent in cell cultures is within a range of 1-10 μg/ml, although it can be toxic to eukaryotic cells at concentrations as low as 1 μg/ml. Puromycin acts quickly and can kill up to 99% of nonresistant cells within 2 days.
Zeocin
[edit | edit source]Zeocin is a glycopeptide antibiotic and one of the phleomycins from Streptomyces verticillus belonging to the bleomycin family of antibiotics. It is a broad-spectrum antibiotic that is effective against most bacteria, filamentous fungi, yeast, plant, and animal cells. It causes cell death by intercalating into DNA and induces double strand breaks of the DNA.
Resistance to zeocin is conferred by the product of the Sh ble gene first isolated from Streptoalloteichus hindustanus. The Sh ble gene product binds the antibiotic in a one to one ratio so it can no longer cause cleavage of DNA.
Useful links
[edit | edit source]- Addgene is a non-profit, global plasmid repository. Addgene facilitates the exchange of genetic material between laboratories by storing plasmids and their associated cloning data.
- The PlasMapper server automatically generates and annotates plasmid maps using only the plasmid DNA sequence as input.
- SnapGene Viewer can be used to view and create richly annotated plasmid maps.
References
[edit | edit source]- Dale, J., Park, S.F., 2004. Molecular genetics of bacteria, 4. ed. Wiley, Chichester.
- Snyder, L., 2013. Molecular genetics of bacteria, 4. ed. ASM Press, Washington, D.C.