MINC/Tools/mni autoreg
What is MNI_AutoReg?
[edit | edit source]In both diagnostic and research applications, the interpretation of magnetic resonance (MR) images of the human brain is facilitated when different data sets can be compared by visual inspection of equivalent anatomical planes. Quantitative analysis with pre-defined atlas templates often requires the initial alignment of atlas and image planes. Unfortunately, the axial planes acquired during separate scanning sessions are often different in their relative position and orientation, and these slices are not coplanar with those in the atlas. A completely automatic method has been developed, based on multi-scale, three dimensional (3D) cross-correlation, to register a given volumetric data set to an average MRI brain (n > 300) aligned with the Talairach stereotaxic coordinate system. Once the data set is resampled by the transformation recovered by the algorithm, atlas slices can be directly super-imposed on the corresponding slices of the resampled volume (see below). The use of such a standardized space also allows the direct comparison, voxel-to-voxel, of two or more data sets brought into stereotaxic space.
A Perl script (mritotal) implements the multi-resolution fitting strategy that has been used to map more than 500 brains into stereotaxic space at the Montreal Neurological Institute. At the heart of this procedure is minctracc, the program that automatically finds the best linear transformation to map one volumetric data set (stored in MINC format, see below) on to another. The program uses optimization over a user selectable number of parameters to identify the best (according to a user-selected objective function) transformation mapping voxel values of the first data set into the second.
Using mni_autoreg
[edit | edit source]The primary scripts for registration are
- mritotal - the script that calls mincblur and minctracc to achieve stereotaxic registration
- mritoself - intra-subject registration
Other scripts or programs installed are:
- autocrop - for extracting and manipulating bounds of a MINC file
- make_model - generate appropriate blurred and subsampled images required for `mritotal'
- make_phantom - generate ellipsoid or rectangular phantom images
- mincbbox - extract bounding box of image
- mincblur - a program for volumetric convolution with a Gaussian blurring kernel
- mincchamfer - compute a Chamfer distance transform of an image
- minctracc - a program to estimate the transformation required for registration
- xfmtool - perform miscellaneous operations on transform (.xfm) files
Some of these ship with manual pages, and all respond to `-help'.
Configuration and Protocol Files
[edit | edit source]mritotal requires two external files to run. These are the configuration file (mritotal.cfg) and protocol file (default is mritotal.default.cfg, but you can specify others using the -protocol option). If you follow all the above instructions, then these files will be correctly customized and installed for your site.
If things go wrong, though, you should know the following:
- The directory where the configuration and protocol file(s) are installed defaults to /usr/local/bic/etc/mni_autoreg.
- The location of the model is given in mritotal.cfg (or in mritotal using the -modeldir option).
- If you find it necessary to make a new protocol for your site's data (for instance, if your MRI data consistently covers considerably more than the brain, but the heuristic used by the default protocol doesn't work - see the mritotal man page for more information), then you should copy and change the default protocol file. For instance, you might want to call the protocol "mysite" - in that case, copy mritotal.default.cfg to mritotal.mysite.cfg, and be sure to edit mritotal.cfg so that it uses the new protocol by default. That is, make sure it has "-protocol mysite" instead of "-protocol default".
What is registration
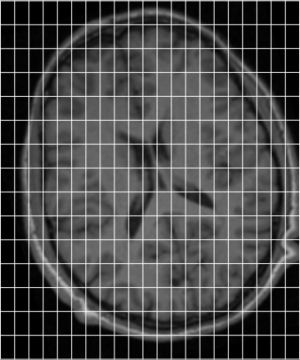
[edit | edit source]In order to get a better understanding of how mni_autoreg or indeed linear registration works, here is a simple example. Suppose we have an image of an individual and wish to align this with a model. These two images are shown below, the individuals image on the left (indiv.mnc) and model (model.mnc) on the right. Note that both of these files have different sampling and voxel sizes as illustrated by the white lines
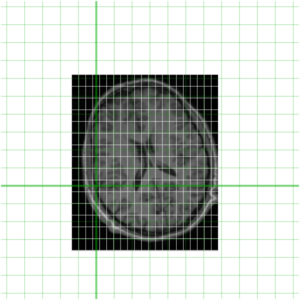
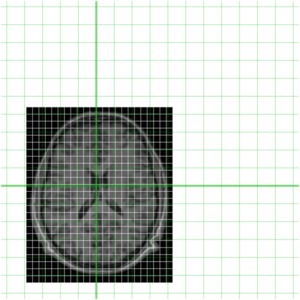
Of course being a MINC file, both of these files represent a sampling of a world space (as defined by starts, steps and direction cosines -- use mincinfo to see this). In order to get an understanding of this, our two files are shown again below with their relative position in world space shown in green.
To get an idea of how they currently overlay I have put them together below
Clearly they are not aligned or "registered" to each other so we have to use a tool such as minctracc or mritoself in order to align them as such:
To do this we can use a command such as this:
minctracc -lsq9 indiv.mnc model.mnc align.xfm
The -lsq9 argument here is instructing minctracc to recover 3 rotations, 3 translations and 3 scales. Note that the output of this step is not an aligned image but an output (linear) transform file which contains a 4x4 tranformation matrix. This consists of a 3x3 affine transformation plus 3 translations should they be needed. An example of one of these files is below:
MNI Transform File Transform_Type = Linear; Linear_Transform = 0.995668319594959 -0.30943698305448 0.0180935790540566 8.9918311534818 0.320977051206147 0.94080272211886 0.00947446085537751 -39.3894632128575 -0.0222220364958048 -0.00403786869719625 1.15379667282104 -33.4353790283203;
A MNI transform file has an implied 4th line consisting of 0,0,0,1 but this data is not stored in the file. To get an idea of what a decomposition of this matrix could be we can use xfm2param:
xfm2param align.xfm
This will produce output like the below:
after parameter extraction -center 0.00000 0.00000 0.00000 -translation 8.99183 -39.38946 -33.43538 -rotation -0.20051 1.10337 17.25793 -scale 1.04280 0.99372 1.15402 -shear 0.02764 -0.00000 -0.00000
So now that we have our transformation we need to resample our original input data. To do this we use mincresample, however we have a few choices to make regarding the sampling of the output file. The first and most commonly used would be as such:
mincresample -like model.mnc -transformation align.xfm source.mnc resampled.mnc
Note that the output file now has the same sampling as the original model.
The next option is as such:
mincresample -tfm_input_sampling -transformation align.xfm source.mnc resampled.mnc
This instructs mincresample to resample the original sampling of the file by the input transformation, note that this will result in a file with direction co-sines.
The final option is to use the original sampling of the individual as such:
mincresample -use_input_sampling -transformation align.xfm source.mnc resampled.mnc
Using this method will keep the original sampling but also the original position of the file meaning you may or may not keep a full "covering" of the file.
It is for this reason that the -like option is the most used approach as it then also allows further processing to be done on the resulting resampled file using voxel based tools such as minccalc and mincmath
Copyright
[edit | edit source]Copyright (C) 1993-2010 Louis Collins and Greg Ward, McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University. Permission to use, copy, modify, and distribute this software and its documentation for any purpose and without fee is hereby granted, provided that the above copyright notice appear in all copies. The authors and McGill University make no representations about the suitability of this software for any purpose. It is provided "as is" without express or implied warranty. The authors are not responsible for any data loss, equipment damage, property loss, or injury to subjects or patients resulting from the use or misuse of this software package. The results of any MNI_AutoReg registration should always be carefully checked by visual inspection in all three spatial dimensions. MNI_AutoReg registrations should never be used in a setting where an incorrect result could injure a patient or subject or lead to an incorrect diagnosis. This package is provided for research use only.
We make no warranty that this package is free of conceptual or programming errors. The fact that this package has been provided to you does not imply or assure any additional support beyond what is provided in this document. Likewise, it does not guarantee that you will be notified of any programming errors that are discovered or that you will be provided with future updated versions of the software. The authors disclaim all liability for direct or indirect damages resulting from your use of Montreal Neurological Institute Automated Linear Registration Package.
We would appreciate that at least the following article be referenced to describe the registration method in all publications of data analyzed using the MNI_AutoReg package:
DL Collins, P Neelin, TM Peters and AC Evans, Automatic 3D Inter-Subject Registration of MR Volumetric Data in Standardized Talairach Space, Journal of Computer Assisted Tomography, 18(2) p192-205, 1994
Bibliography
[edit | edit source]D. L. Collins, P. Neelin, T. M. Peters and A. C. Evans, ``Automatic 3D Inter-Subject Registration of MR Volumetric Data in Standardized Talairach Space, Journal of Computer Assisted Tomography, 18(2) pp192-205, 1994.