Structural Biochemistry/Nucleic Acid/DNA
Introduction
[edit | edit source]


What is DNA? DNA is a long chain of linear polymers containing deoxyribose sugars and their covalently bonded bases known as nucleic acids. One of the major functions of the DNA is storage of the genetic information. In DNA a sequence of three bases, which is called a codon, is responsible for the encoding of a single amino acid. The amino acid is added to a growing protein during the process of translation. These nucleic acid polymers encode for the all of the materials an organism needs to live in the form of genes. Genes are small blocks of DNA that tell the cell which proteins it should create. The type of genes that a given cell receives depends entirely on the parent cells. Genes are passed on from generation to generation as a way of ensuring an organism's survival genetically.
DNA stands for deoxyribonucleic Acid. The prefix "deoxy" distinguishes DNA from its close relative RNA (ribonucleic acid). The prefix indicates that, unlike Ribose, Deoxyribose does not contain a hydroxyl group at the 2' carbon replacing it with a single Hydrogen atom. The absence of this Hydroxyl group is fundamental in determining the way in which DNA is able to condense itself within the nucleus of a cell.
DNA is a nucleic acid which is capable of duplicating itself via the enzyme known as DNA polymerase. Each of the four bases on DNA, Adenine (A), Cytosine (C), Guanine (G), and Thymine (T) is bonded covalently to a deoxyribose sugar. The four nucleotide units in DNA are called deoxyadenylate, deoxyguanylate, deoxycytidylate, and thymidylate. The nucleotide includes the nucleoside, a nitrogenous base bonded to a deoxyribose or ribose group. The four nucleosides in DNA are deoxyadenosine, deoxyguanosine, deoxycytidine, and thymide. By the joining one or more phosphate groups to a nucleoside through ester linkages, a nucleotide is formed.
The deoxyribose sugars form the structural backbone for DNA via a phosphodiester bond between the 3' carbon of one nucleotide and the 5' carbon of the next. When DNA is not self-replicating it exists in the cell as a double stranded helical molecule with the strands lined up anti-parallel to each other. That is to say if the orientation of one strand is 3' to 5' the other strand would be oriented 5' to 3'. The bases of each strand bind very specifically, A binds with T and C binds with G no other combination exists at least in DNA. The bases are bound to one another internally via hydrogen bonds with the phosphodiester bond backbone oriented to face outward. It is here that the missing 2' hydroxyl group plays an important role in DNA. It is the absence of this group that allows DNA to form its conventional double helix structure. RNA which does have a hydroxyl group at the 2' carbon is unable to obtain this same helical structure. The modern double helix structure of DNA was first proposed by Watson and Crick, and the functions of DNA were demonstrated in a series of experiments which will be discussed in the next few sections.
Why DNA? It is significant to note the reasons why DNA is the primary method through which all cells pass along genetic information. That is to say why has evolution favored a DNA world over an RNA world given that the two molecules are so similar structurally? These reasons involve chemical stability, energy needed to form and break chemical bonds, and the availability of enzymes to perform this task. The primary reason involves the relative stability of the two molecules. DNA is more chemically stable than RNA because it lacks the hydroxyl group on the 2' carbon. In RNA there are two possible OH groups that the molecule can form a phosphodiester bond between, which means that RNA is not forced into the same rigid structure as its deoxy counterpart. Additionally the deoxyribose sugar in DNA is much less reactive than the ribose sugar in RNA. Simply put C-H groups are significantly less reactive than C-OH (hydroxyl) groups. This difference also explains why RNA is not very stable in alkaline conditions, and DNA is. The base in alkaline condition does the same thing as the -OH group at the C2 position. Furthermore, double-strand DNA has relatively small grooves where damaging enzymes can't attach, making it more difficult for them to 'attack' the DNA. Double-stranded RNA, on the other hand, has much larger grooves, and therefore, it is more subject to being broken down by enzymes. The connection between the strands of double-stranded DNA is tighter than double-stranded RNA. In other words, it's much easier to unzip double-stranded RNA than it is to unzip double-stranded DNA. Overall, the breakdown and reform of RNA can be carried out faster and requires less energy than the breakdown and reform of DNA. It is essential to the organism's survival and well-being that its genetic material is encoded into something that is more stable and resistant to changes. In addition, the sequence of DNA and its physical conformation seems to play a part in DNA's selection as well. Another point that helps elucidate DNA's prevalence as the primary storage of genetic information is the availability of the enzyme that breaks down DNA. The body actively destroys foreign nucleases, which are enzymes that cleave DNA. This is only one of the many ways DNA is protected against damage. The body can actually recognize foreign DNA and destroy it, while leaving its own DNA intact.
Hyperchromic Effect Another unique feature of DNA in its double stranded form is the hyperchromic effect, which describes the decreasing absorbance of UV electromagnetic radiation of double helix strands as compared to the non-helical conformation of the molecule. The hydrogen bonding between complementary DNA strands as a result of sugar stacking in the helical conformation causes the aromatic rings to become increasingly stable and thus absorb less UV radiation. This ultimately decreases the amount of UV absorption by 40%. As the temperature is increased these hydrogen bonds dissolve and the helical structure begins to unwind. In this unwound form the aromatic rings are free to absorb much more UV radiation.
Properties of DNA
[edit | edit source]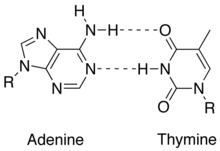

1. Consists of 2 strands (anti-parallel and complementary): DNA has two polynucleotide chains that twist around a helical axis in opposite direction.
2. It is made up of deoxyribose sugar, a phosphate backbone on the exterior, and nucleic acid bases in the interior.
3. Bases are perpendicular to the helix axis that separated by 3.4 Angstroms.
4. Strands are held together by hydrogen bonds an other various intermolecular forces that form a double helix. The base pairing involves 2 hydrogen bonds for A - T and 3 hydrogen bonds for C - G -see in images to the right
5. Backbone consists of alternating sugars and phosphates, where phosphodiester linkages form the covalent backbone of the DNA.The direction of DNA goes from 5' phosphate group to 3' hydroxide group.
6. Repeats every 10 bases
7. Weak forces stabilize DNA because of the hydrophobic effects and VanDerWaals.
8. DNA chain is 20 Angstroms wide (2 nm)
9. One nucleotide unit is 3.3 Angstroms long (0.33 nm)
Primary Structure
[edit | edit source]DNA is made of two polynucleotide chains (strands) which run in opposite directions around the common axis. As a result, DNA has a double helical structure. Each polynucleotide chain of DNA consists of monomer units. A monomer unit consists of three main components that are a sugar, a phosphate, and a nitrogenous base. The sugar used in the DNA monomer unit is deoxyribose (it lacks an oxygen atom on the second Carbon in the furanose ring). There are also four possible nitrogen containing bases which can be used in the monomer unit of the DNA. Those bases are adenine (A), guanine (G), cytosine (C), and thymine (T). Adenine and guanine are purine derivatives, while cytosine and thymine are pyrimidine derivatives. Polymeric chain forms as a result of joining nucleosides (the sugar which is covalently bonded to the nitrogen containing base) through the phosphodiester linkage. Polymeric chain is a single strand of the DNA molecule. Two strands run in opposite directions to form double helix. The forces that keep those strands together are hydrogen bond, hydrophobic interactions, van de Waal force, and charge-charge interactions. The H-bonds form between base pairs of the antiparallel strands. The base in the first strand forms an H-bond only with a specific base in the second strand. Those two bases form a base-pair (H-bond interaction that keeps strands together and form double helical structure). The base –pairs are: adenine-thymine (A-T), cytosine-guanine (C-G). Such interaction gives us the hint that nitrogen-containing bases are located inside of the DNA double helical structure, while sugars and phosphates are located outside of the double helical structure. The hydrophobic bases are inside the double helix of DNA. The bases, located inside the double helix, are stacked one on the top of another. Stacking bases interact with each other through the Van der Waals force. Even though the van de Waal forces are week, sumation of those forces can be substantial. The distance between two neighboring bases that are perpendicular to the main axis is 3.4 A˚. DNA structure is repetitive. There are ten bases per turn, so every base has a 36° angle of rotation. The diameter of the double helix is approximately 20 A˚. The hydrophobic effect stabilizes the double helix. The structural variation in DNA is due to the different deoxyribose conformations, rotation about the contiguous bonds in the phosphodeoxyribose backbone, and free rotation about the C-1'- N (glycosyl bond).
The technique of southern blotting is often used to uncover the DNA sequence of a sample. The technique is named after Edwin Southern.
DNA Manipulation Techniques
[edit | edit source]When it comes to exploring genes and genomes, it depends on the technical tools that are used. The five important DNA manipulation techniques are:
1.Restriction Endonucleases - also known as restriction enzymes
The restriction of enzymes split the DNA into specific fragments. By having the DNA split into different pieces, it allows the manipulation of DNA segments.
2. Blotting Technique
To separate and characterize DNA, the Southern blotting technique is used. This technique is similar to the Western blot, except that Southern blotting is used for DNA and not RNA. This technique identifies a specific sequence of DNA by electrophoresis through an agarose gel. The DNA is separated by placing the large fragments on top and the small fragments at the bottom. Next, the DNA is transfer into the nitrocellulose sheet. Then a 32-p labeled DNA probe that is complementary to the sequence, is added to hybridize the fragments. Finally, a autoradiography film is use to view the fragment containing the sequence.
3. DNA Sequencing
By using the DNA sequencing technique, a precise nucleotide sequence of a DNA molecule can be determined. The key to DNA sequencing is the generation of DNA fragments whose length depends on the last base of the sequence. Even though there are different alternative methods, they all perform the same procedure on the four reaction mixtures.
A. Chain termination DNA Sequencing
A primer is always needed. To produce fragments, the addition of 2', 3'-dideoxy analog of a dNTP is added to each of the four mixtures. It will stop the sequence at that N-dideoxy. The types of dNTP that can be use are dATP, TTP, dCTP, dGTP. In the end, new DNA strands are separated to electrophoresis.
B. Fluorescence Detection of Bases
Fluorescent tag is used into each of the four chain-terminating dideoxy nucleotides at different wavelengths. It is an effective method because no radioactive reagents are used and large sequences of bases can be determined. The fragments get separated by having the mixture passed through high voltage. Then, the fragments are detected by their fluorescence, which the base sequence is based on the color sequence.
C. Top-down (Shotgun) Method of Genome Sequencing
The top-down method and the shotgun method are similar, the main difference is that the top-down requires a detailed map of the clones. The Shotgun randomly sequences large clones to match them computationally.
D. Microarrays(Green chips)
Using microarrays is useful when it comes to studying the expression of a large number of genes. The microarray is created by using either oligonucleotides or cDNA. Based on the fluorescent intensity, red or green marks will appear. If it is red, it means no fluorescence is present, known as gene induction. If it is green, fluorescence is expressed, known as gene repression.
4. DNA Synthesis
To synthesize DNA, a solid-phase method is used. The solid-phase synthesis is carried out by the phosphite triester method. In this process only one nucleotide is added in each group. The first step that takes place is the binding of the first nucleotide. Another nucleotide is added and activated and reacts with the 3' -phosphoramididte containing DMT. A deoxyribonucleoside 3' -phosphoramidite with DMT and βCE is attached because it has the ability to synthesize any DNA. It is also a basic nucleotide that is modified and protected. Then, the molecule gets oxidized to oxidized the phosphate group. In the end, the DMT is removed by addition of dichloroacetic acid. Overall, the desired product remains insoluble and it is release at the end.
5. Polymerase Chain Reaction (PCR)
PCR is a technique used that allows to amplify DNA sequence between two nucleotides. If the DNA sequence is known, millions of copies of that sequence can be obtained by using this technique. To carried out PCR, a DNA template, a precursor, and two complementary primers are needed. What makes the PCR unique is that the temperature is constantly changing within the three different stages and that the stages get repeated 25 times. The three stages are:
1. Denaturing - DNA gets denature from a double strand (parent DNA molecule) to two single strands by heating thesolution at 94°C.
2. Annealing - After letting the solution cooled, two synthetic oligonucleotide primers are added at the end of the 3' end of target strand, and at the 3' end of complementary strand. This process is done when the temperature is between 50°C - 60°C.
3. Polymerization - Addition of thermostable DNA polymerase to catalyze 5' to 3' DNA synthesis at 72°C.
Structural Variation
[edit | edit source]Structural Variation occurs due to the different deoxyribose conformations, free rotation about the C-1, and rotation about the closest bond in phosphodeoxyribose backbones.
There are secondary structures when it comes to DNA which are forms A, B, and Z. A Form: 1. Right handed 2. Glycosyl bond conformation is ANTI 3. Needs 11 base pairs per helical turn 4. Size of diameter is about 26 angstroms 5. Sugar pucker conformation is at the C-3' endo.
B Form: 1. Like the A form, the B form is right handed. 2. Glycosyl bond formation is ANTI 3. Needs 10.5 base pairs per helical turn 4. Size of diameter is about 20 angstroms 5. Sugar pucker conformation is at the C-2' endo
Z Form: 1. Unlike the A and B form, the obvious difference is that the Z form is left handed. 2. Glycosyl bond formation consists of two components: pyrimidines and purines. ANTI (for pyrimidines) and SYN (for purines) 3. Needs 12 base pairs per helical turn 4. Size of diameter is about 18 angstroms 5. Sugar pucker conformation is at the C-2' endo (for pyrimidines) and C-3' endo (for purines)
DNA libraries
[edit | edit source]A DNA library is a collection of cloned DNA fragments in a cloning vector that can be searched for a DNA of interest. If the goal is to isolate particular gene sequences, two types of library are useful.
Genomic DNA libraries
[edit | edit source]A genomic DNA library is made from the genomic DNA of an organism. For example, a mouse genomic library could be made by digesting mouse nuclear DNA with a restriction nuclease to produce a large number of different DNA fragments but all with identical cohesive ends. The DNA fragments would then be ligated into linearized plasmid vector molecules or into a suitable virus vector. This library would contain all of the nuclear DNA sequences of the mouse and could be searched for any particular mouse gene of interest. Each clone in the library is called a genomic DNA clone. Not every genomic DNA clone would contain a complete gene since in many cases the restriction enzyme will have cut at least once within the gene. Thus some clones will contain only a part of a gene.
cDNA Library
[edit | edit source]A cDNA library is a library of mRNAs. It is made from introns and exons and a cDNA library is made to be able to isolate the genes/the final version of the gene.
A cDNA library i used to screen for colonies. If looking for a gene, you can screen the colonies, use the collection of plasmids, transform the bacteria, and use a probe. You can also use Southern Hybridization. By using an oligonucleotide that is complementary to the gene you are looking for, and that will eventually tell you which colonies of bacteria will have the DNA that corresponds with the mRNA in the plasmids.
How to make a cDNA library:
1. Isolate mRNA from the cell.
2. Use reverse transcriptase and dNTPss so that from the original mRNA, a DNA copy can be created.
3. RNA is easier to degrade than DNA so put in alkali solution to degrade mRNA.
4. Use DNA polymerase to complete the template.
Ultimately, you end up with double stranded DNA, one of which is identical to the mRNA. After doing this all for mRNA, you can clone it in the plasmids. The collection of plasmids will include all of the mRNA but in the form of DNA.[1]
Flow of Genetic Information
[edit | edit source]- Genetic information storage: genome
- Replication: DNA --> DNA
- Transcription: DNA --> RNA
- Translation: RNA --> Proteins
References
[edit | edit source]- ↑ Viadiu, Hector. "Making a cDNA Library." UCSD. Lecture. November 2012.
Sources
[edit | edit source]http://clem.mscd.edu/~churchcy/BIO3600/bio3600_images/phosphodiester_bonds.htm http://www.science-projects.com/hyperChromic1.htm
Berg , Jeremy . Biochemistry . 7. New York : W.H Freeman and Company , 2012. Print.
| This page or section is an undeveloped draft or outline. You can help to develop the work, or you can ask for assistance in the project room. |
Berg, Jeremy, Tymoczko J., Stryer, L.(2012). Protein Composition and Structure.Biochemistry(7th Edition). W.H. Freeman and Company. ISBN1-4292-2936-5
Hames, David. Hooper, Nigel. Biochemistry. Third edition. New York. Taylor and Francis Groups. 2005.
