Methods and Concepts in the Life Sciences/Microscopy
Microscopy[edit | edit source]
Microscopy is the technical field of using microscopes to view objects and areas of objects that cannot be seen with the naked eye (objects that are not within the resolution range of the normal eye). There are three well-known branches of microscopy: optical, electron, and scanning probe microscopy.
Bright-field microscopy[edit | edit source]

Bright-field microscopy is the simplest of all the optical microscopy illumination techniques. Sample illumination is transmitted (i.e., illuminated from below and observed from above) white light and contrast in the sample is caused by absorbance of some of the transmitted light in dense areas of the sample.
Dark field microscopy[edit | edit source]

Dark field microscopy describes an illumination technique used to enhance the contrast in unstained samples. It works by excluding the unscattered beam from the image. This produces the classic appearance of a dark background with bright objects on it.
The light path through a dark field microscope is as follows:
- Light enters the microscope for illumination of the sample.
- A specially sized disc, the patch stop blocks some light from the light source, leaving an outer ring of illumination. A wide phase annulus can also be reasonably substituted at low magnification.
- The condenser lens focuses the light towards the sample.
- The light enters the sample. Most is directly transmitted, while some is scattered from the sample.
- The scattered light enters the objective lens, while the directly transmitted light simply misses the lens and is not collected due to a direct illumination block.
- Only the scattered light goes on to produce the image, while the directly transmitted light is omitted.
Dark field microscopy is a very simple yet effective technique and well suited for uses involving live and unstained biological samples, such as a smear from a tissue culture or individual, water-borne, single-celled organisms. Considering the simplicity of the setup, the quality of images obtained from this technique is impressive.
The main limitation of dark field microscopy is the low light levels seen in the final image. This means the sample must be very strongly illuminated, which can cause damage to the sample. Dark field microscopy techniques are almost entirely free of artifacts, due to the nature of the process. However, the interpretation of dark field images must be done with great care, as common dark features of bright field microscopy images may be invisible, and vice versa.
Phase contrast microscopy[edit | edit source]

Phase contrast is a widely used technique that shows differences in refractive index as difference in contrast.
The basic principle to make phase changes visible in phase contrast microscopy is to separate the illuminating background light from the specimen scattered light, which make up the foreground details, and to manipulate these differently.
The ring shaped illuminating light that passes the condenser annulus is focused on the specimen by the condenser. Some of the illuminating light is scattered by the specimen. The remaining light is unaffected by the specimen and forms the background light. When observing an unstained biological specimen, the scattered light is weak and typically phase shifted by -90° relative to the background light. This entails that the foreground and the background nearly have the same intensity, resulting in a low image contrast (a).
In a phase contrast microscope, the image contrast is improved in two steps. The background light is phase shifted -90° by passing it through a phase shift ring. This eliminates the phase difference between the background and the scattered light, leading to an increased intensity difference between foreground and background (b). To further increase contrast, the background is dimmed by a gray filter ring (c). Some of the scattered light will be phase shifted and dimmed by the rings. However, the background light is affected to a much greater extent, which creates the phase contrast effect.

The above describes negative phase contrast. In its (more common) positive form, the background light is instead phase shifted by +90°. The background light will thus be 180° out of phase relative to the scattered light. This means that the scattered light will be subtracted from the background light in (b) to form an image where the foreground is darker than the background.
Differential interference contrast (DIC) microscopy[edit | edit source]

Differential interference contrast (DIC) microscopy is an optical microscopy illumination technique used to enhance the contrast in unstained, transparent samples. A relatively complex lighting scheme produces an image with the object appearing black to white on a grey background. This image is similar to that obtained by phase contrast microscopy but without the bright diffraction halo.
The system consists of a special prism (Nomarski prism, Wollaston prism) in the condenser that splits light in an ordinary and an extraordinary beam. The spatial difference between the two beams is minimal (less than the maximum resolution of the objective). After passage through the specimen, the beams are reunited by a similar prism in the objective.
In a homogeneous specimen, there is no difference between the two beams, and no contrast is being generated. However, near a refractive boundary (say a nucleus within the cytoplasm), the difference between the ordinary and the extraordinary beam will generate a relief in the image. Differential interference contrast requires a polarized light source to function; two polarizing filters have to be fitted in the light path, one below the condenser (the polarizer), and the other above the objective (the analyzer).
The light path[edit | edit source]
- Unpolarised light enters the microscope and is polarised at 45°.
- The polarised light enters the first Nomarski-modified Wollaston prism and is separated into two rays polarised at 90° to each other, the sampling and reference rays.
- The two rays are focused by the condenser for passage through the sample. These two rays are focused so they will pass through two adjacent points in the sample, around 0.2 μm apart.
- The rays travel through adjacent areas of the sample, separated by the shear. The separation is normally similar to the resolution of the microscope. They will experience different optical path lengths where the areas differ in refractive index or thickness. This causes a change in phase of one ray relative to the other due to the delay experienced by the wave in the more optically dense material. The passage of many pairs of rays through pairs of adjacent points in the sample (and their absorbance, refraction and scattering by the sample) means an image of the sample will now be carried by both the 0° and 90° polarised light. These, if looked at individually, would be bright field images of the sample, slightly offset from each other. The light also carries information about the image invisible to the human eye, the phase of the light. The different polarisations prevent interference between these two images at this point.
- The rays travel through the objective lens and are focused for the second Nomarski-modified Wollaston prism.
- The second prism recombines the two rays into one polarised at 135°. The combination of the rays leads to interference, brightening or darkening the image at that point according to the optical path difference.

The image[edit | edit source]
The image has the appearance of a three-dimensional object under very oblique illumination, causing strong light and dark shadows on the corresponding faces. The direction of apparent illumination is defined by the orientation of the Wollaston prisms.
As explained above, the image is generated from two identical bright field images being overlaid slightly offset from each other (typically around 0.2 μm), and the subsequent interference due to phase difference converting changes in phase (and so optical path length) to a visible change in darkness. This interference may be either constructive or destructive, giving rise to the characteristic appearance of three dimensions.
The typical phase difference giving rise to the interference is very small, very rarely being larger than 90° (a quarter of the wavelength). This is due to the similarity of refractive index of most samples and the media they are in: for example, a cell in water only has a refractive index difference of around 0.05. This small phase difference is important for the correct function of DIC, since if the phase difference at the joint between two substances is too large then the phase difference could reach 180° (half a wavelength), resulting in complete destructive interference and an anomalous dark region; if the phase difference reached 360° (a full wavelength), it would produce complete constructive interference, creating an anomalous bright region.
DIC has strong advantages in uses involving live and unstained biological samples, such as a smear from a tissue culture or individual water borne single-celled organisms. Its resolution and clarity in conditions such as this are unrivaled among standard optical microscopy techniques.
The main limitation of DIC is its requirement for a transparent sample of fairly similar refractive index to its surroundings. DIC is unsuitable (in biology) for thick samples, such as tissue slices, and highly pigmented cells. DIC is also unsuitable for most non biological uses because of its dependence on polarisation, which many physical samples would affect.
Fluorescence microscopy[edit | edit source]
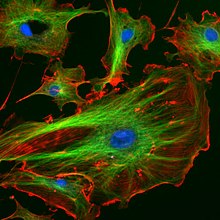
A fluorescence microscope is an optical microscope that uses fluorescence and phosphorescence instead of, or in addition to, reflection and absorption.
The specimen is illuminated with light of a specific wavelength (or wavelengths) which is absorbed by the fluorophores, causing them to emit light of longer wavelengths. The illumination light is separated from the much weaker emitted fluorescence through the use of a spectral emission filter. Typical components of a fluorescence microscope are a light source (xenon arc lamp or mercury-vapor lamp are common; more advanced forms are high-power LEDs and lasers), the excitation filter, the dichroic mirror (or dichroic beamsplitter), and the emission filter. The filters and the dichroic are chosen to match the spectral excitation and emission characteristics of the fluorophore used to label the specimen. In this manner, the distribution of a single fluorophore is imaged at a time. Multi-color images of several types of fluorophores must be composed by combining several single-color images.

Most fluorescence microscopes in use are epifluorescence microscopes, where excitation of the fluorophore and detection of the fluorescence are done through the same light path (i.e. through the objective). These microscopes are widely used in biology and are the basis for more advanced microscope designs, such as the confocal microscope and the total internal reflection fluorescence microscope (TIRF).
Deconvolution[edit | edit source]
Fluorescence microscopy is a powerful technique to show specifically labeled structures within a complex environment and to provide three-dimensional information of biological structures. However, this information is blurred by the fact that, upon illumination, all fluorescently labeled structures emit light, irrespective of whether they are in focus or not. So an image of a certain structure is always blurred by the contribution of light from structures that are out of focus. This phenomenon results in a loss of contrast especially when using objectives with a high resolving power, typically oil immersion objectives with a high numerical aperture.
However, blurring is not caused by random processes, such as light scattering, but can be well defined by the optical properties of the image formation in the microscope imaging system. If one considers a small fluorescent light source (essentially a bright spot), light coming from this spot spreads out further from our perspective as the spot becomes more out of focus. Under ideal conditions, this produces an "hourglass" shape of this point source in the third (axial) dimension. This shape is called the point spread function (PSF) of the microscope imaging system. Since any fluorescence image is made up of a large number of such small fluorescent light sources, the image is said to be "convolved by the point spread function".
Knowing this point spread function means that it is possible to reverse this process to a certain extent by computer-based methods commonly known as deconvolution microscopy. There are various algorithms available for 2D or 3D deconvolution. They can be roughly classified in nonrestorative and restorative methods. While the nonrestorative methods can improve contrast by removing out-of-focus light from focal planes, only the restorative methods can actually reassign light to its proper place of origin. Processing fluorescent images in this manner can be an advantage over directly acquiring images without out-of-focus light, such as images from confocal microscopy, because light signals otherwise eliminated become useful information. For 3D deconvolution, one typically provides a series of images taken from different focal planes (called a Z-stack) plus the knowledge of the PSF, which can be derived either experimentally or theoretically from knowing all contributing parameters of the microscope.
Confocal microscopy[edit | edit source]
Confocal microscopy is an optical imaging technique for increasing optical resolution and contrast of a micrograph by means of adding a spatial pinhole placed at the confocal plane of the lens to eliminate out-of-focus light. It enables the reconstruction of three-dimensional structures from the obtained images.
In a conventional (i.e., wide-field) fluorescence microscope, the entire specimen is flooded evenly in light from a light source. All parts of the specimen in the optical path are excited at the same time and the resulting fluorescence is detected by the microscope's photodetector or camera including a large unfocused background part. In contrast, a confocal microscope uses point illumination and a pinhole in an optically conjugate plane in front of the detector to eliminate out-of-focus signal - the name "confocal" stems from this configuration. As only light produced by fluorescence very close to the focal plane can be detected, the image's optical resolution, particularly in the sample depth direction, is much better than that of wide-field microscopes. However, as much of the light from sample fluorescence is blocked at the pinhole, this increased resolution is at the cost of decreased signal intensity, so long exposures are often required.

As only one point in the sample is illuminated at a time, 2D or 3D imaging requires scanning over a regular raster (i.e., a rectangular pattern of parallel scanning lines) in the specimen. The achievable thickness of the focal plane is defined mostly by the wavelength of the used light divided by the numerical aperture of the objective lens, but also by the optical properties of the specimen. The thin optical sectioning possible makes these types of microscopes particularly good at 3D imaging and surface profiling of samples.
Spinning-disk microscope[edit | edit source]
Spinning-disk confocal microscopes use a series of moving pinholes on a disc to scan spot of light. Since a series of pinholes scans an area in parallel each pinhole is allowed to hover over a specific area for a longer amount of time thereby reducing the excitation energy needed to illuminate a sample when compared to laser scanning microscopes. Decreased excitation energy reduces photo-toxicity and photo-bleaching of a sample often making it the preferred system for imaging live cells or organisms.
Fluorescence correlation spectroscopy (FCS)[edit | edit source]

The typical FCS setup consists of a laser line, which is reflected into a microscope objective by a dichroic mirror. The laser beam is focused in the sample, which contains fluorescent molecules in such high dilution, that only a few are within the focal spot (usually 1–100 molecules in one fL). When the particles cross the focal volume, they fluoresce. This light is collected by the same objective and, because it is red-shifted with respect to the excitation light, it passes the dichroic mirror reaching a detector. The resulting electronic signal can be stored either directly as an intensity versus time trace to be analyzed at a later point, or computed to generate the autocorrelation directly. The FCS curve by itself only represents a time-spectrum. Conclusions on physical phenomena have to be extracted from there with appropriate models. The parameters of interest are found after fitting the autocorrelation curve to modeled functional forms. When an appropriate model is known, FCS can be used to obtain quantitative information such as diffusion coefficients, hydrodynamic radii, average concentrations and kinetic chemical reaction rates.
Fluorescence-lifetime imaging microscopy (FLIM)[edit | edit source]
Fluorescence-lifetime imaging microscopy (FLIM) is an imaging technique for producing an image based on the differences in the decay rate of the fluorescence from a fluorescent sample.
This has the advantage of minimizing the effect of photon scattering in thick layers of sample. Fluorescence-lifetime imaging yields images with the intensity of each pixel determined by the fluorescence lifetime, which allows one to view contrast between materials with different fluorescence decay rates (even if those materials fluoresce at exactly the same wavelength), and also produces images which show changes in other decay pathways, such as in FRET imaging.
Total internal reflection fluorescence microscopy (TIRFM)[edit | edit source]

A total internal reflection fluorescence microscope (TIRFM) is a type of microscope with which a thin region of a specimen, usually less than 200 nm can be observed.
In cell and molecular biology, a large number of molecular events in cellular surfaces such as cell adhesion, binding of cells by hormones, secretion of neurotransmitters, and membrane dynamics have been studied with conventional fluorescence microscopes. However, fluorophores that are bound to the specimen surface and those in the surrounding medium exist in an equilibrium state. When these molecules are excited and detected with a conventional fluorescence microscope, the resulting fluorescence from those fluorophores bound to the surface is often overwhelmed by the background fluorescence due to the much larger population of non-bound molecules.
A TIRFM uses an evanescent wave to selectively illuminate and excite fluorophores in a restricted region of the specimen immediately adjacent to the glass-water interface. The evanescent wave is generated only when the incident light is totally internally reflected at the glass-water interface. The evanescent electromagnetic field decays exponentially from the interface, and thus penetrates to a depth of only approximately 100 nm into the sample medium. Thus, the TIRFM enables a selective visualization of surface regions such as the basal plasma membrane (which are about 7.5 nm thick) of cells as shown in the figure above. Note, however, that the region visualised is at least a few hundred nanometers wide, so the cytoplasmic zone immediately beneath the plasma membrane is necessarily visualised in addition to the plasma membrane during TIRF microscopy. The selective visualisation of the plasma membrane renders the features and events on the plasma membrane in living cells with high axial resolution.
TIRF can also be used to observe the fluorescence of a single molecule, making it an important tool of biophysics and quantitative biology.
Fluorescence recovery after photobleaching (FRAP) and Fluorescence Loss in Photobleaching (FLIP)[edit | edit source]
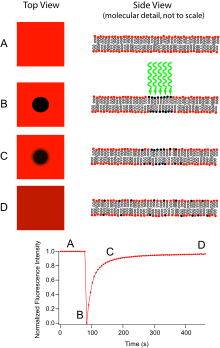
Fluorescence recovery after photobleaching (FRAP) denotes an optical technique capable of quantifying the two dimensional lateral diffusion of a molecularly thin film containing fluorescently labeled probes, or to examine single cells. This technique is very useful in biological studies of cell membrane diffusion and protein binding. In addition, surface deposition of a fluorescing phospholipid bilayer (or monolayer) allows the characterization of hydrophilic (or hydrophobic) surfaces in terms of surface structure and free energy.
The basic apparatus comprises an optical microscope, a light source and some fluorescent probe. The technique begins by saving a background image of the sample before photobleaching. Next, the light source is focused onto a small patch of the viewable area. The fluorophores in this region receive high intensity illumination which causes their fluorescence lifetime to quickly elapse. Now the image in the microscope is that of a uniformly fluorescent field with a noticeable dark spot. As Brownian motion proceeds, the still-fluorescing probes will diffuse throughout the sample and replace the non-fluorescent probes in the bleached region. This diffusion proceeds in an ordered fashion, analytically determinable from the diffusion equation.
Fluorescence Loss in Photobleaching (FLIP) is closely associated with FRAP. The major difference between these two microscopy techniques is that FRAP involves the study of a cell’s ability to recover after a single photobleaching event whereas FLIP involves the study of how the loss of fluorescence spreads throughout the cell after multiple photobleaching events. This difference in purpose also leads to a difference in what parts of the cell are observed. In FRAP, the area that is actually photobleached is the area of interest. Conversely, in FLIP, the region of interest is just outside the region that is being photobleached. Another important difference is that in FRAP, there is a single photobleaching event and a recovery period to observe how well fluorophores move back to the bleached site. However, in FLIP, multiple photobleaching events occur to prevent the return of unbleached fluorophores to the bleaching region. Like FLIP, FRAP is used in the study of continuity of membranous organelles. FLIP and FRAP are often used together to determine the mobility of GFP-tagged proteins. FLIP can also be used to measure the molecular transfer between regions of a cell regardless of the rate of movement. This allows for a more comprehensive analysis of protein trafficking within a cell. This differs from FRAP which is primarily useful for determining mobility of proteins in regions local to the photobleaching only.
Super-resolution microscopy[edit | edit source]
Super-resolution microscopy is a form of light microscopy. Due to the diffraction of light, the resolution of conventional light microscopy is limited as stated by Ernst Abbe in 1873. A good approximation of the resolution attainable is the full width at half maximum (FWHM) of the point spread function, and a precise widefield microscope with high numerical aperture and visible light usually reaches a resolution of ~250 nm.
4Pi[edit | edit source]
A 4Pi microscope is a laser scanning fluorescence microscope with an improved axial resolution. The typical value of 500–700 nm can be improved to 100–150 nm, which corresponds to an almost spherical focal spot with 5–7 times less volume than that of standard confocal microscopy.
The improvement in resolution is achieved by using two opposing objective lenses both of which focused to the same geometrical location. Also the difference in optical path length through each of the two objective lenses is carefully aligned to be minimal. By this, molecules residing in the common focal area of both objectives can be illuminated coherently from both sides and also the reflected or emitted light can be collected coherently, i.e. coherent superposition of emitted light on the detector is possible. The solid angle that is used for illumination and detection is increased and approaches the ideal case. In this case the sample is illuminated and detected from all sides simultaneously.
Structured illumination microscopy (SIM)[edit | edit source]

Structured-illumination microscopy relies on both specific microscopy protocols and extensive software analysis post-exposure. The main concept of SI is to illuminate a sample with patterned light and increase the resolution by measuring the fringes in the Moiré pattern (from the interference of the illumination pattern and the sample). Otherwise-unobservable sample information can be deduced from the fringes and computationally restored.
SI enhances spatial resolution by collecting information from frequency space outside the observable region. This process is done in reciprocal space: The Fourier transform (FT) of an SI image contains superimposed additional information from different areas of reciprocal space; with several frames with the illumination shifted by some phase, it is possible to computationally separate and reconstruct the FT image, which has much more resolution information. The reverse FT returns the reconstructed image to a super-resolution image.
Stimulated emission depletion (STED)[edit | edit source]

STED (Stimulated Emission Depletion microscopy) uses two laser pulses, the excitation pulse for excitation of the fluorophores to their fluorescent state and the STED pulse for the de-excitation of fluorophores by means of stimulated emission. In practice, the excitation laser pulse is first applied whereby a STED pulse soon follows (But STED without pulses using continuous wave lasers is also used). Furthermore, the STED pulse is modified in such a way that it features a zero-intensity spot, which coincides with the excitation focal spot. Due to the non-linear dependence of the stimulated emission rate on the intensity of the STED beam, all the fluorophores around the focal excitation spot will be in their off state (the ground state of the fluorophores). By scanning this focal spot one retrieves the image.
Normal fluorescence occurs by exciting an electron from the ground state into an excited electronic state of a different fundamental energy level (S0 goes to S1) which, after relaxing back to the ground state (of s1), emits a photon by dropping from S1 to a vibrational energy level on S0. STED interrupts this process before the photon is released. The excited electron is forced to relax into a higher vibration state than the fluorescence transition would enter, causing the photon to be released to be red-shifted. Because the electron is going to a higher vibrational state, the energy difference of the two states is lower than the normal fluorescence difference. This lowering of energy raises the wavelength, and causes the photon to be shifted farther into the red end of the spectrum. This shift differentiates the two types of photons, and allows the stimulated photon to be ignored.
To force this alternative emission to occur, an incident photon must strike the fluorophore. This need to be struck by an incident photon has two implications for STED. First, the number of incident photons directly impacts the efficiency of this emission, and, secondly, with sufficiently large numbers of photons fluorescence can be completely suppressed. To achieve the large number of incident photons needed to suppress fluorescence, the laser used to generate the photons must be of a high intensity. Unfortunately, this high intensity laser can lead to the issue of photobleaching the fluorophore.
STORM, PALM and FPALM[edit | edit source]
Stochastic optical reconstruction microscopy (STORM), photo activated localization microscopy (PALM) and fluorescence photo-activation localization microscopy (FPALM) are super-resolution imaging techniques that utilize sequential activation and time-resolved localization of photoswitchable fluorophores to create high resolution images. During imaging, only an optically resolvable subset of fluorophores is activated to a fluorescent state at any given moment, such that the position of each fluorophore can be determined with high precision by finding the centroid position of the single-molecule images of particular fluorophores. The fluorophore is subsequently deactivated, and another subset is activated and imaged. Iteration of this process allows numerous fluorophores to be localized and a super-resolution image to be constructed from the image data. These three methods were published independently during a short period of time and their principle is identical. STORM was originally described using Cy5 and Cy3 dyes attached to nucleic acids or proteins, while PALM and FPALM was described using photoswitchable fluorescent proteins. In principle any photoswitchable fluorophore can be used.
Useful links[edit | edit source]
References
- Ishikawa-Ankerhold, H.C., Ankerhold, R., Drummen, G.P.C., 2012. Advanced fluorescence microscopy techniques--FRAP, FLIP, FLAP, FRET and FLIM. Molecules 17, 4047–132. doi:10.3390/molecules17044047
- Leung, B.O., Chou, K.C., 2011. Review of super-resolution fluorescence microscopy for biology. Appl. Spectrosc. 65, 967–80. doi:10.1366/11-06398
- Lichtman, J.W., Conchello, J.-A., 2005. Fluorescence microscopy. Nat. Methods 2, 910–9. doi:10.1038/nmeth817
- Petty, H.R., 2007. Fluorescence Microscopy : Established and Emerging Methods, Experimental Strategies, and Applications in Immunology 709, 687–709. doi:10.1002/jemt