Data Mining Algorithms In R/Classification/Decision Trees
Introduction
[edit | edit source]The philosophy of operation of any algorithm based on decision trees is quite simple. In fact, although sometimes containing important differences in the way to do this or that step, any algorithm of this category is based on the strategy of divide and conquer. In general, this philosophy is based on the successive division of the problem into several subproblems with a smaller number of dimensions, until a solution for each of the simpler problems can be found. Based on this principle, the classifiers based on decision trees try to find ways to divide the universe into successively more subgroups (creating nodes containing the respective tests) until each addressing only one class or until one of the classes shows a clear majority do not justifying further divisions, generating in this situation a leaf containing the class majority. Obviously, the classification is only to follow the path dictated by the successive test placed along the tree until it found a leaf containing the class to assign to the new example.
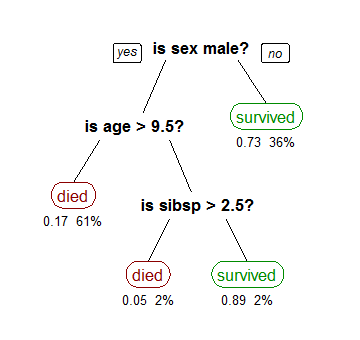
Although the basic philosophy of all the classifiers based on decision trees is identical, there are many possibilities for its construction. Among all the key points in the selection of an algorithm to build decision trees some of them should be highlighted for their importance:
- Criteria for the choice of feature to be used in each node
- How to calculate the partition of the set of examples
- When you decide that a node is a leaf
- What is the criterion to select the class to assign to each leaf
Some important advantages can be pointed to the decision trees, including:
- Can be applied to any type of data
- The final structure of the classifier is quite simple and can be stored and handled in a graceful manner
- Handles very efficiently conditional information, subdividing the space into sub-spaces that are handled individually
- Reveal normally robust and insensitive to misclassification in the training set
- The resulting trees are usually quite understandable and can be easily used to obtain a better understanding of the phenomenon in question. This is perhaps the most important of all the advantages listed
Algorithm
[edit | edit source]The basic algorithm for decision tree is the greedy algorithm that constructs decision trees in a top-down recursive divide-and-conquer manner. We usually employ greedy strategies because they are efficient and easy to implement, but they usually lead to sub-optimal models. A bottom-up approach could also be used. The top-down decision tree algorithm is given in Algorithm 1. It is a recursive divide-and-conquer algorithm. It takes a subset of data D as input and evaluate all possible splits (Lines 4 to 11). The best split decision (Line 12), i.e. the split with the highest information gain, is chosen to partition the data in two subsets (divide-and-conquer) and the method is called recursively (Lines 14 and 15). The algorithm stops when the stop conditions are met (Line 1 to 3).
Stopping Criteria
[edit | edit source]A number of stopping conditions can be used to stop the recursive process. The algorithm stops when any one of the conditions is true:
- All the samples belong to the same class, i.e. have the same label since the sample is already "pure"
- Stop if most of the points are already of the same class. This is a generalization of the first approach, with some error threshold
- There are no remaining attributes on which the samples may be further partitioned
- There are no samples for the branch test attribute
Attribute Selection
[edit | edit source]We now need an objective criteria for judging how good a split is. The information gain measure is used to select the test attribute at each node in the tree. The attribute with the highest information gain (or greatest entropy reduction) is chosen as the test attribute for the current node. This attribute minimizes the information needed to classify the samples in the resulting partitions.
Entropy, in general, measures the amount of disorder or uncertainty in a system. In the classification setting, higher entropy (i.e., more disorder) corresponds to a sample that has a mixed collection of labels. Lower entropy corresponds to a case where we have mostly pure partitions. In information theory, the entropy of a sample D is defined as follows:
where is the probability of a data point in D being labeled with class , and k is the number of classes. can be estimated directly from the data as follows:
We can also define the weighted entropy of a decision/split as follows:
where D has been partitioned into and due to some split decision. Finally, we can define the information gain for a given split as:
In other words, Gain is the expected reduction in entropy caused by knowing the value of an attribute.
Rpart
[edit | edit source]The rpart package found in the R tool can be used for classification by decision trees and can also be used to generate regression trees. Recursive partitioning is a fundamental tool in data mining. It helps us explore the structure of a set of data, while developing easy to visualize decision rules for predicting a categorical (classification tree) or continuous (regression tree) outcome. The rpart programs build classification or regression models of a very general structure using a two stage procedure; the resulting models can be represented as binary trees. The tree is built by the following process: first the single variable is found which best splits the data into two groups ('best' will be defined later). The data is separated, and then this process is applied separately to each sub-group, and so on recursively until the subgroups either reach a minimum size (5 for this data) or until no improvement can be made. The resultant model is, with certainty, too complex, and the question arises as it does with all stepwise procedures of when to stop. The second stage of the procedure consists of using cross-validation to trim back the full tree.
Grow the Tree
[edit | edit source]To grow a tree, use
rpart (formula, data=, method=, control=)
where:
| formula | is in the format: outcome ~ predictor1+predictor2+predictor3+ect. |
| data= | specifies the dataframe |
| method= | "class" for a classification tree "anova" for a regression tree |
| control= | optional parameters for controlling tree growth. For example, control=rpart.control(minsplit=30, cp=0.001) requires that the minimum number of observations in a node be 30 before attempting a split and that a split must decrease the overall lack of fit by a factor of 0.001 (cost complexity factor) before being attempted. |
Visualization and Examples
[edit | edit source]printcp
[edit | edit source]The command printcp displays the cp table for fitted rpart object. Prints a table of optimal prunings based on a complexity parameter.
Usage
printcp(object, digits=getOption("digits") - 2)
where object is an rpart object and digits is the number of digits of numbers to print.
Example:
fit <- rpart(Price ~ HP, car.test.frame)
printcp(fit)
Output
Regression tree:
rpart(formula = Price ~ HP, data = car.test.frame)
Variables actually used in tree construction:
[1] HP
Root node error: 983551497/60 = 16392525
n= 60
CP nsplit rel error xerror xstd
1 0.41417 0 1.00000 1.03808 0.21528
2 0.12304 1 0.58583 0.71817 0.15575
3 0.01000 2 0.46279 0.62650 0.11675
plotcp
[edit | edit source]The command plotcp gives a visual representation of the cross-validation results in an rpart object. The set of possible cost-complexity prunings of a tree from a nested set. For the geometric means of the intervals of values of cp for which a pruning is optimal, a cross-validation has (usually) been done in the initial construction by rpart. The cptable in the fit contains the mean and standard deviation of the errors in the cross-validated prediction against each of the geometric means, and these are plotted by this function. A good choice of cp for pruning is often the leftmost value for which the mean lies below the horizontal line.
Usage
plotcp(object, minline = TRUE, lty = 3, col = 1, upper = c("size", "splits", "none"), args)
Where object is an rpart object, minline is whether a horizontal line is drawn 1SE above the minimum of the curve, lty is the line type for this line, col is the colour for this line upper what is plotted on the top axis: the size of the tree (the number of leaves), the number of splits or nothing and args are the arguments to be passed to or from other methods.
Example:
fit <- rpart(Kyphosis ~ Age + Number + Start, method="class", data=kyphosis)
plotcp(fit)
rsq.rpart
[edit | edit source]Plot approximate r-squared versus the number of splits and relative error for different splits versus the number of splits (two plots).
Usage
rsq.rpart(object)
Where object is an rpart object.
Example:
fit <- rpart(Mileage ~ Weight, car.test.frame)
rsq.rpart(fit)
Prints an rpart object.
Usage
print(object, minlength=0, spaces=2, cp, digits= getOption("digits"), args)
where object is an rpart object, minlength controls the abbreviation of labels, spaces is the number of spaces to indent nodes of increasing depth, digits is the number of digits of numbers to print, cp prune all nodes with a complexity less than cp from the printout and args are the arguments to be passed to or from other methods.
Example:
fit <- rpart(Kyphosis ~ Age + Number + Start, method="class", data=kyphosis)
print(fit)
n= 81
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 81 17 absent (0.7901235 0.2098765)
2) Start>=8.5 62 6 absent (0.9032258 0.0967742)
4) Start>=14.5 29 0 absent (1.0000000 0.0000000) *
5) Start< 14.5 33 6 absent (0.8181818 0.1818182)
10) Age< 55 12 0 absent (1.0000000 0.0000000) *
11) Age>=55 21 6 absent (0.7142857 0.2857143)
22) Age>=111 14 2 absent (0.8571429 0.1428571) *
23) Age< 111 7 3 present (0.4285714 0.5714286) *
3) Start< 8.5 19 8 present (0.4210526 0.5789474) *
summary
[edit | edit source]Returns a detailed listing of a fitted rpart object.
Usage
summary(object, cp=0, digits=getOption("digits"), file, args)
Where object is an rpart object, digits is the number of significant digits to be used in the result, cp trim nodes with a complexity of less than cp from the listing, file write the output to a given file name and args are the arguments to be passed to or from other methods.
Example:
fit <- rpart(Mileage ~ Weight, car.test.frame)
summary(fit)
Call:
rpart(formula = Mileage ~ Weight, data = car.test.frame)
n= 60
CP nsplit rel error xerror xstd
1 0.59534912 0 1.0000000 1.0294527 0.17907324
2 0.13452819 1 0.4046509 0.6261647 0.10545991
3 0.01282843 2 0.2701227 0.4746041 0.08567822
4 0.01000000 3 0.2572943 0.4884699 0.08551818
Node number 1: 60 observations, complexity param=0.5953491
mean=24.58333, MSE=22.57639
left son=2 (45 obs) right son=3 (15 obs)
Primary splits:
Weight < 2567.5 to the right, improve=0.5953491, (0 missing)
Node number 2: 45 observations, complexity param=0.1345282
mean=22.46667, MSE=8.026667
left son=4 (22 obs) right son=5 (23 obs)
Primary splits:
Weight < 3087.5 to the right, improve=0.5045118, (0 missing)
Node number 3: 15 observations
mean=30.93333, MSE=12.46222
Node number 4: 22 observations
mean=20.40909, MSE=2.78719
Node number 5: 23 observations, complexity param=0.01282843
mean=24.43478, MSE=5.115312
left son=10 (15 obs) right son=11 (8 obs)
Primary splits:
Weight < 2747.5 to the right, improve=0.1476996, (0 missing)
Node number 10: 15 observations
mean=23.8, MSE=4.026667
Node number 11: 8 observations
mean=25.625, MSE=4.984375
plot (and text)
[edit | edit source]Plots an rpart object on the current graphics device as a decision tree. The function text label the decision tree plot.
Usage
plot(object, uniform=FALSE, branch=1, compress=FALSE, nspace, margin=0, minbranch=.3, args)
Where object is an rpart object, uniform if TRUE uniform vertical spacing of the nodes is used, branch controls the shape of the branches from parent to child node, compress if FALSE, the leaf nodes will be at the horizontal plot coordinates of 1:nleaves, if TRUE, the routine attempts a more compact arrangement of the tree, nspace is the amount of extra space between a node with children and a leaf, margin is an extra percentage of white space to leave around the borders of the tree, minbranch set the minimum length for a branch to minbranch times the average branch length and args are the arguments to be passed to or from other methods.
Example:
fit <- rpart(Price ~ Mileage + Type + Country, cu.summary)
plot(fit, compress=TRUE)
text(fit, use.n=TRUE)
post
[edit | edit source]Create a PostScript presentation plot of an rpart object.
Usage
plot(object, uniform=FALSE, branch=1, compress=FALSE, nspace, margin=0, minbranch=.3, args)
Object is an rpart object, uniform if TRUE uniform vertical spacing of the nodes is used, branch controls the shape of the branches from parent to child node, compress if FALSE, the leaf nodes will be at the horizontal plot coordinates of 1:nleaves, if TRUE, the routine attempts a more compact arrangement of the tree, nspace is the amount of extra space between a node with children and a leaf, margin is an extra percentage of white space to leave around the borders of the tree, minbranch set the minimum length for a branch to minbranch times the average branch length and args are the arguments to be passed to or from other methods.
Example:
fit <- rpart(Mileage ~ Weight, car.test.frame)
post(fit, file = "", title="Classification Tree for Wikibook")
post(fit, file = "c:/output.ps", title = " Classification Tree for Wikibook")
Case Study
[edit | edit source]Scenario and Input data
[edit | edit source]Consider the relational database in the table below, whose schema is composed of attributes Play, Outlook, Temperature, Humidity and Windy. A decision tree allows predicting the values of the attribute Play, given that we know the values for attributes like Outlook, Humidity and Windy.
| weather | Temperature | Humidity | Wind | Golf play |
|---|---|---|---|---|
| fine | hot | high | none | no |
| fine | hot | high | few | no |
| cloud | hot | high | none | yes |
| rain | warm | high | none | yes |
| rain | cold | mediam | none | yes |
| rain | cold | mediam | few | no |
| cloud | cold | mediam | few | yes |
| fine | warm | heigh | none | no |
| fine | cold | mediam | none | yes |
| rain | warm | mediam | none | yes |
| fine | warm | mediam | few | yes |
| cloud | warm | high | few | yes |
| cloud | hot | mediam | none | yes |
| rain | warm | high | few | no |
Importing data into R is simple. From a comma delimited text (CSV) file whose the first row contains variable names we can use the command below:
play_base <- read.table("path_to_the_file/play.csv", header=TRUE, sep=",")
We can use the command print(play_base) or just play_base to see the loaded table and the command "summary(play_base)" to see a detailed listing of the rpart object:
Play Outlook Temperature Humidity Windy
no :3 overcast:2 cool:5 high :4 false:7
yes:7 rainy :4 hot :2 normal:6 true :3
sunny :4 mild:3
Execution and Output
[edit | edit source]After we have loaded the data we need to build the decision tree. The "Play" attribute is the outcome that will the predicted. We can use the command below:
fit <- rpart(Play ~ Outlook + Temperature + Humidity + Wind, method="class", data=play_base,
control=rpart.control(minsplit=1))
We can use the command summary(fit) to see a detailed listing of the loaded decision tree and the command print(fit) to see the decision tree:
1) root 10 3 yes (0.3000000 0.7000000)
2) Temperature= mild 3 1 no (0.6666667 0.3333333)
4) Outlook= sunny 2 0 no (1.0000000 0.0000000) *
5) Outlook= overcast 1 0 yes (0.0000000 1.0000000) *
3) Temperature= cool, hot 7 1 yes (0.1428571 0.8571429)
6) Windy= true 1 0 no (1.0000000 0.0000000) *
7) Windy= false 6 0 yes (0.0000000 1.0000000) *
The commands below plots an rpart object on the current graphics device as a decision tree:
plot(fit, uniform=TRUE, main="Decision Tree - Play?")
text(fit, use.n=TRUE, all=TRUE, cex=.8)
Analysis
[edit | edit source]The building of a decision tree starts with a description of a problem which should specify the variables, actions and logical sequence for a decision-making. In a decision tree, a process leads to one or more conditions that can be brought to an action or other conditions, until all conditions determine a particular action, once built you can have a graphical view of decision-making.
The decision tree generated to solve the problem, the sequence of steps described determines and the weather conditions, verify if it is a good choice to play or not to play. For instance, in the sequence of conditions (temperature = mild) -> (Outlook = overcast) -> play = yes, whereas in the sequence (temperature = cold) -> (Windy = true) -> play = no. This shows that a decision tree is a great tool for making decisions. Thus, this method of classification may become an excellent tool for obtaining information, which often organizations do not know they have, and which are extremely important to the tactical and management level.
References
[edit | edit source]Jiawei Han. Data Mining: Concepts and Techniques. Morgan Kaufmann Publishers, 2001.
Mohammed J. Zaki and Wagner Meira Jr.. Fundamentals of Data Mining Algorithms. Cambridge University Press, 2010.
Terry M. Therneau, Elizabeth J. Atkinson and Mayo Foundation. An Introduction to Recursive Partitioning Using the RPART Routines, 1997.
R Language Definition - [1]
An Introduction to R - [2]
Quick-R - [3]
Terry M Therneau and Beth Atkinson. Package ‘rpart’, 2009.








