A-level Applied Science/The Role of the Pathology Service/Microbiology
Microbiology laboratories receive swabs, faeces, urine, blood, sputum, medical equipment, as well as possible infected tissue. They culture this to check for any pathogenic microbes.
Pathogenic microbes may be bacteria, fungi, viruses or other parasites.
Viruses are difficult to culture because they require living cells to reproduce. Virology is therefore done in larger medical laboratories.
Bacteria, fungi and other non-viral parasites can grow independently of other living organisms if they are provided with the correct nutrients and growth conditions. Culturing the organism in this way will assist in identifying it and studying possible therapies against it.
Infectious diseases
[edit | edit source]An infectious disease is a clinically evident disease of humans or animals that damages or injures the host so as to impair host function, and results from the presence and activity of a pathogenic microbial agent, including viruses, bacteria, fungi, protozoa, multicellular parasites, and aberrant proteins known as prions. Transmission of an infectious disease may occur through several pathways; including through contact with infected individuals, by water, food, airborne inhalation, or through vector-borne spread.[1]
A contagious disease (also called a communicable disease) is an infectious disease that is capable of being transmitted from one person or species to another.[2] Contagious diseases are often spread through direct contact with an individual, contact with the bodily fluids of infected individuals, or with objects that the infected individual has contaminated.
The term infectivity describes the ability of an organism to enter, survive and multiply in the host, while the infectiousness of a disease indicates the comparative ease with which the disease is transmitted to other hosts.[3] An infection however, is not synonymous with an infectious disease; as an infection may not cause clinical symptoms or impair host function.[1]
Overview
[edit | edit source]Among the almost infinite varieties of microorganisms, relatively few cause a disease in an otherwise normal or healthy individual.[4] These highly virulent microorganisms are classified as primary pathogens. Organisms, which cause an infectious disease in a host with depressed resistance, are classified as opportunistic pathogens. Opportunistic infectious disease may be caused by organisms that are ordinarily in contact with the host, such as bacteria or fungi in the colon or in the upper respiratory tract; following an injury, whether mechanical (such as a open Bone fracture|fracture); or by a disease with immunosuppressive activity (such as measles or malaria, or one induced by cytotoxic chemotherapy). Primary pathogens may also cause more explosive disease in a host with depressed resistance.
Most primary pathogens are inhabitants of, and pathogens for, humans only. Opportunistic pathogens, in contrast, may cause disease in many species of mammals. Some exceptions are tetanus, anthrax and rabies which may inhabit and cause disease in many animal species including humans.
Agents and vectors
[edit | edit source]Infectious disease requires an agent and a mode of transmission (or vector). A good example is malaria, which is caused by Plasmodial parasites, chiefly Plasmodium falciparum but does not affect humans unless the vector, the Anopheles mosquito, is around to introduce the parasite into the human bloodstream.
The vector does not have to be biological. Many infectious diseases are transmitted by droplets which enter the airway (e.g. common cold and tuberculosis).
Clearance and immunity
[edit | edit source]Infection with most pathogens does not result in death of the host and the offending organism is ultimately cleared after the symptoms of the disease have waned.[4] This process requires immune mechanisms to kill or inactivate the inoculum of the pathogen. Specific acquired immunity against infectious diseases may be mediated by antibodies and/or T lymphocytes. Immunity mediated by these two factors may be manifested by:
- a direct effect upon a pathogen, such as antibody-initiated complement-dependent bacteriolysis, opsonoisation, phagocytosis and killing, as occurs for some bacteria,
- neutralization of viruses so that these organisms cannot enter cells,
- or by T lymphocytes which will kill a cell parasitized by a micro organism.
The immune response to a micro organism often causes symptoms such as a high fever and inflammation, and has the potential to be more devastating than direct damage caused by a microbe.
Resistance to infection (immunity) may be acquired following a disease, by asymptomatic carriage of the pathogen, by harbouring an organism with a similar structure (cross reacting), or by vaccination. Knowledge of the protective antigens and specific acquired host immune factors is more complete for primary pathogens than for opportunistic pathogens. Immune resistance to an infectious disease requires a critical level of either antigen-specific antibodies and/or T cells when the host encounters the pathogen. Some individuals develop natural serum antibodies to the surface polysaccharides of some agents although they have had little or no contact with the agent, these natural antibodies confer specific protection to adults and are transmitted to newborns.
The work of an infectiologist
[edit | edit source]Doctors who specialise in the medical treatment of infectious disease are called infectiologists or infectious disease specialists. Generally, infections are initially diagnosed by primary care physicians or internal medicine specialists. For example, an "uncomplicated" pneumonia will generally be treated by the internist or the pulmonologist (lung physician).
The services of the infectious disease team are called for when:
- The disease has not been definitively diagnosed after an initial workup
- The patient is immunocompromised (for example, in AIDS or after chemotherapy);
- The infectious agent is of an uncommon nature (e.g. tropical diseases);
- The disease has not responded to first line antibiotics;
- The disease might be dangerous to other patients, and the patient might have to be isolated.
The work of the infectiologist therefore entails working with patients and doctors on one hand and laboratory scientists and immunologists on the other hand.
Mortality from infectious diseases
[edit | edit source]The relationship between virulence and transmission is complex, and has important consequences for the long term evolution of a pathogen.
If a disease is rapidly fatal, the host may die before the microbe can get passed along to another host. However, this cost may be overwhelmed by the short term benefit of higher infectiousness if transmission is linked to virulence, as it is for instance in the case of cholera (the explosive diarrhoea aids the bacterium in finding new hosts) or many respiratory infections (sneezing, coughing etc. create infectious aerosols).
The classification of infectious disease
[edit | edit source]Epidemiology is another important tool used to study disease in a population. For infectious diseases it helps to determine if a disease outbreak is sporadic (occasional occurrence), endemic (regular cases often occurring in a region), epidemic (an unusually high number of cases in a region), or pandemic (a global epidemic).
Collecting samples
[edit | edit source]Before starting treatment, the physician will collect a sample from a suspected infected compartment: a blood sample when bacteria possibly have invaded the bloodstream, a sputum sample, a urine sample, etc. These samples are transferred to the microbiology lab, which looks at the sample under the microscope, and tries to culture the bacteria. This can help in the diagnosis.
Serology
[edit | edit source]Serum is the yellow watery part of blood that is left after blood has been allowed to clot and all blood cells have been removed. This is most easily done by centrifugation which packs the more dense blood cells and platelets to the bottom of the centrifuge tube, leaving the liquid serum fraction resting above the packed cells. Serologists receive serum samples to look for evidence of diseases such as hepatitis or HIV.
Certain agents cannot be microbiologically cultured, for example Treponema pallidum and most viruses. The first serological markers were developed to diagnose syphilis (the Wassermann test, later replaced by the VDRL and TPHA tests). Serology involves detecting the antibodies against an infectious agent in the patient's blood. In immunocompromised patients (e.g. AIDS), serology can be troublesome, because the antibody reaction is blunted.
A more recent development is direct detection of viral proteins and/or DNA in blood or secretions. This can be done by PCR (polymerase chain reaction), involving the amplification of viral DNA and its subsequent detection with anti-DNA probes.
Aseptic technique
[edit | edit source]Aseptic technique is the name given to the procedures used by microbiologists to prevent microbial contamination of themselves, which may result in infection, contamination of the environment they are working in, and contamination of the specimen they are working on, which is especially important when a pure culture is desired. It is used whenever specimens are to be transferred between media, for example, when subculturing. Such a procedure, using a flame sterilisation method, might occur as follows:

- A person would assemble the closed tube or flask from which — and the closed tube or flask to which — the specimen is to be transferred, an inoculating loop, and a flame source, all on a clean, preferably microbe-free surface with some overhead protection from airborne microbes.
- The person would start the flame, and move the end of the inoculating loop, in a slow back-and-forth motion, through the top of the blue part of the flame. The person would not allow the loop to touch anything except the specimen itself, until the entire procedure is finished.
- Preparing to execute the specimen transfer, the person would hold both of the tubes or flasks in one hand, probably the opposite of the writing hand. The person would then open the tube or flask containing the specimen source and briefly hold the top of it in the flame, to kill unwanted microbes.
- Quickly, so as to minimize the possible time for contamination of the specimen in the source tube or flask, the person would use the inoculating loop with their writing hand to retrieve the specimen, and then sterilize the top of the tube or flask again before immediately closing it.
- Keeping in mind that the specimen on the inoculating loop could be contaminated every of time it is exposed, the person would repeat the previous step identically with the tube or flask in which the specimen is to be deposited; however, the person would be depositing the sample into the tube or flask.
Students of microbiology are taught the principles of aseptic technique by means of hands-on laboratory experience. Practice is critical to learning how to handle the laboratory tools without contaminating them.
Microbiological cultures
[edit | edit source]Diagnosis is initially by medical history and physical examination, and imaging (such as X-rays), but the principal tool in infectious disease is the microbiological culture. In a culture, a growth medium is provided for a particular agent. After inoculation of a specimen of diseased fluid or tissue onto the medium, it is determined whether bacterial growth occurs. This works for a number of bacteria, for example Staphylococcus or Streptococcus.

A microbiological culture, or microbial culture, is a method of growing a microbial organism to determine what it is, its abundance in the sample being tested, or both. It is one of the primary diagnostic methods of microbiology. It is often used a tool to determine the cause of infectious disease by letting the agent multiply (reproduce) in predetermined media in laboratory.
The most common method of microbiological culture uses Petri dishes with a layer of agar-based growth medium in them to grow bacterial cultures. This is generally done inside of an incubator. Another method is liquid culture, where the bacteria are grown suspended in a liquid nutrient medium. Bottles of liquid culture are often placed in shakers in order to introduce oxygen to the liquid and maintaining the uniformity of the culture.
The term culture can also, though infrequently and informally, be used as a synonym for tissue culture, which involves the growth of cells or tissues explanted from a multi-cellular organism.
A sputum culture is a test to detect and identify bacteria or fungi that are infecting the lungs or breathing passages. Sputum is a thick fluid produced in the lungs and in the airways leading to the lungs. A sample of sputum is placed in a container with substances that promote the growth of bacteria or fungi. If no bacteria or fungi grow, the culture is negative. If organisms that can cause infection (pathogenic organisms) grow, the culture is positive. The type of bacterium or fungus will be identified with a microscope or by chemical tests.
If bacteria or fungi that can cause infection grow in the culture, other tests may be done to determine which antibiotic will be most effective in treating the infection. This is called susceptibility or sensitivity testing.
This test is done on a sample of sputum that is usually collected by coughing. For people who cannot cough deeply enough to produce a sample, a suction tube or needle may be inserted in the airway to collect the sputum.
In a hospital setting, a sputum culture is most commonly ordered if a patient has a pneumonia. The Infectious Disease Society of America recommends that sputum cultures be done in pneumonia requiring hospitalization, while the American College of Chest Physicians does not. The reasons for the discrepancy is this: normal, healthy lungs have bacteria, and sputum cultures collect both normal bacteria and the bacteria causing disease. It is difficult to tell which bacteria is causing the disease in this setting.
Antibiograms
[edit | edit source]The role of antibiograms
[edit | edit source]In clinical practice, antibiotics are most frequently prescribed on the basis of general guidelines and knowledge about sensitivity: e.g. uncomplicated urinary tract infections can be treated with a first generation quinolone, etc. This is because Escherichia coli is the most likely causative pathogen, and it is known to be sensitive to quinolone treatment. Infections that are not acquired in the hospital, are called "community acquired" infections.
However, many bacteria are known to be resistant to several classes of antibiotics, and treatment is not so straightforward. This is especially the case in vulnerable patients, such as patients in the intensive care unit. When these patients develop a "hospital-acquired" (or "nosocomial") pneumonia, more hardy bacteria like Pseudomonas aeruginosa are potentially involved. Treatment is then generally started on the basis of surveillance data about the local pathogens probably involved. This first treatment, based on statistical information about former patients, and aimed at a large group of potentially involved microbes, is called "empirical treatment".
When a culture has proven to be positive, the sensitivity (or, conversely, the antibiotic resistance) of an agent can be determined by exposing it to test doses of antibiotic. This way, the microbiologist determines how sensitive the target bacterium is to a certain antibiotic. This is usually reported as being: Sensitive, Intermediate or Resistant. The antibiogram can then be used to determine optimal therapy for the patient. This can reduce the use of broad-spectrum antibiotics and lead to a decrease in antibiotic resistance.

An antibiogram is the result of a laboratory testing for the sensitivity of an isolated bacterial strain to different antibiotics. It is by definition an in vitro-sensitivity test.
Antibiogram methods
[edit | edit source]Once a culture is established, there are two possible ways to get an antibiogram:
- a semi-quantitative way based on diffusion (Kirby-Bauer method); small discs containing different antibiotics, or impregnated paper discs, are dropped in different zones of the culture in the Petri dish. The antibiotic will diffuse in the area surrounding each tablet, and a disc of bacterial lysis will become visible. Since the concentration of the antibiotic was the highest at the centre, and the lowest at the edge of this zone, the diameter is suggestive for the Minimum Inhibitory Concentration (conversion of the diameter in millimeter to the MIC, in µg/ml, is based on known linear regression curves).
- a quantitative way based on dilution: a dilution series of antibiotics is established (this is a series of reaction vials with progressively lower concentrations of antibiotic substance). The last vial in which no bacteria grow contains the antibiotic at the Minimal Inhibiting Concentration.
Once the MIC is calculated, it can be compared to know values for a given bacterium and antibiotic: e.g. a MIC > 0,06 µg/ml may be interpreted as a penicillin-resistant Streptococcus pneumoniae. Such information may be useful to the clinician, who can change the empirical treatment, to a more custom-tailored treatment that is directed only at the causative bacterium.
Health and safety
[edit | edit source]Waste materials from this department are likely to be highly infectious. Their safe disposal is very important and microbiology laboratories will have rigorous disposal procedures.
Practical work
[edit | edit source]Candidates could be given a range of ‘unknown’ cultures which they need to characterise by looking at colony morphologies, growth characteristics, media requirements, and further characterise by performing Gram staining.
Candidates could investigate the antimicrobial activity of liquid soaps, toothpastes or similar household products which make antibacterial claims.
If possible a range of different bacteria or fungi could be investigated and links made to reasons behind the different efficacies of antibiotics.
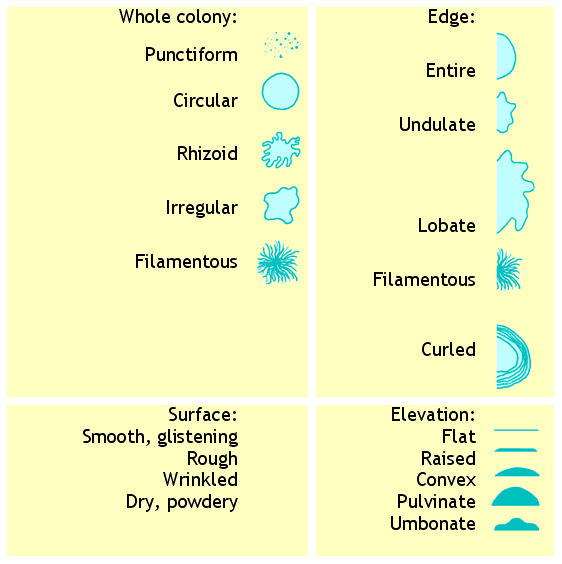
Bacterial colony shape
[edit | edit source]The shape (morphology) of bacterial colonies on an agar plate is a valuable aid to identifying the bacteria.
Bacterial cell shape
[edit | edit source]The shape (morphology) of bacterial cells under the microscope is a valuable aid to identifying the bacteria.
Gram staining
[edit | edit source]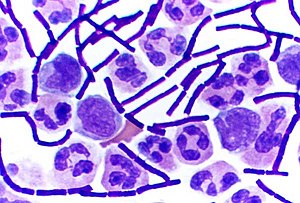

Gram staining (or Gram's method) is an empirical method of differentiating bacterial species into two large groups based on the chemical and physical properties of their cell walls.
The method is named after the inventor, the Danish scientist Hans Christian Gram (1853–1938), who developed the technique in 1884.
Uses
[edit | edit source]Gram stains are performed on body fluid or biopsy when infection is suspected. It yields results much quicker than culture, and is especially important when infection would make an important difference in the patient's treatment and prognosis; examples are cerebrospinal fluid for meningitis and synovial fluid for septic arthritis. It necessitates the 24 hour staffing of microbiological laboratories in hospitals.
Mechanism
[edit | edit source]Gram-positive bacteria have a thick mesh-like cell wall made of peptidoglycan which is capable of retaining the violet dye/iodine complex. Gram-negative bacteria have a thin cell wall made of a layer of peptidoglycan. In addition to an inner membrane, they also have an outer membrane which contains lipids, and is separated from the cell wall by the periplasmic space.
The decolourising mixture causes dehydration of the multilayered peptidoglycan in the Gram-positive cell wall, thus decreasing the space between the molecules and causing the cell wall to trap the crystal violet-iodine complex within the cell. But in Gram-negative bacteria, the decolourising mixture acts as a lipid solvent and dissolves the outer membrane of the Gram-negative cell wall. The thin layer of peptidoglycan is unable to retain the crystal violet-iodine complex and the Gram-negative cell is decolourised. The decolourisation step is the crucial one, and requires some degree of skill, as Gram-positivity is not an all-or-none phenomenon.
As a rule of thumb (which has exceptions), Gram-negative bacteria are more dangerous as disease organisms, because their outer membrane is often hidden by a capsule or slime layer which hides the antigens of the cell and so acts as "camouflage" - the human body recognises a foreign body by its antigens; if they are hidden, it becomes harder for the body to detect the invader. Often the presence of a capsule will increase the virulence of a pathogen. Additionally, Gram-negative bacteria have lipopolysaccharide in their outer membrane. Lipopolysaccharide is an endotoxin which increases the severity of inflammation. This inflammation may be so severe that septic shock may occur. Gram-positive infections are generally less severe because the human body does not contain peptidoglycan, and in fact the human body produces an enzyme called lysozyme which attacks the open peptidoglycan layer of Gram-positive bacteria. Gram-positive bacteria are also much more susceptible to beta-lactam antibiotics, such as penicillin.
Procedure
[edit | edit source]- First, an inoculum is taken from a culture using an inoculation loop and put on a slide and then allowed to air dry. If the culture is solid, it is diluted by adding a drop of water or sterile saline on the slide and mixing with the loop. It is important here to take a very small inoculum so that the end result is a sparse single layer of bacteria. It is a common mistake for beginners to put far too much inoculum at this step.
- The specimen is heat-fixed by passing the slide, inoculum side up, through a Bunsen flame 1-2 times, without allowing the slide to become hot to the touch. This prevents the bacteria from being washed away later and it also kills the bacteria
- A basic dye, crystal violet or gentian violet, is used to stain the slide. This dye is taken up by both Gram-positive and Gram-negative bacteria. Allow to stain for 1 minute. The slide should look purple to the unaided eye, and if examined microscopically at this point both Gram-positive and Gram-negative bacteria are purple. Lugol can also be used instead of crystal violet.
- Rinse off with water for a maximum of 5 seconds.
- Add iodine (Gram's iodine) solution (1% iodine, 2% potassium iodide in water) for 1 minute. This acts as a mordant and fixes the dye.
- Rinse with water.
- Apply 95% ethanol or a mixture of propanone and alcohol several times until no more colour appears to come from the sample. This washes away all the unbound basic dye, (usually crystal violet) and leaves Gram-positive organisms stained purple and Gram-negative organisms unstained (colourless).
- Rinse with water immediately to prevent over-decolourisation.
- Apply a suitable counterstain. Opinions vary as to the best choice but suitable stains include safranin or fuchsin. This stain is taken up by both Gram-positive and Gram-negative organisms, but does not alter the colour of Gram-positive organism much, as they are already purple. It does, however, make the Gram-negative organisms pinkish-red.
- Blot gently and allow to dry. Do not smear.
Interpretation
[edit | edit source]Inspect the slide under a microscope
- Gram-positive organisms will appear blue-black or purple.
- Gram-negative organisms will appear red or pink.
Organisms that cannot reliably be differentiated by this staining technique are said to be Gram-variable
See also
[edit | edit source]- w:Infection
- w:Microbiology
- w:List of infectious diseases
- w:Copenhagen Consensus
- Important publications in infectious disease
- w:Big killer
- Staining
References
[edit | edit source]- ↑ a b "Infectious disease." McGraw-Hill Encyclopedia of Science and Technology. The McGraw-Hill Companies, Inc., 2005.
- ↑ Dorland's Illustrated Medical Dictionary 2004 WB Saunders.
- ↑ Glossary of Notifiable Conditions Washington State Department of Health
- ↑ a b This section incorperates public domain materials included in the text: Medical Microbiology Fourth Edition: Chapter 8 (1996) . Baron, Samuel MD. The University of Texas Medical Branch at Galveston.
- "MICROBIOLOGY: Classroom Assignment #2", a Portable Document Format computer file.
- H. Krauss, A. Weber, M. Appel, B. Enders, A. v. Graevenitz, H. D. Isenberg, H. G. Schiefer, W. Slenczka, H. Zahner: Zoonoses. Infectious Diseases Transmissible from Animals to Humans. 3rd Edition, 456 pages. ASM Press. American Society for Microbiology, Washington DC., USA. 2003. ISBN 1-55581-236-8
- Gram HC. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpreparäten. Fortschr Med 1884;2:185-89.
- Bergey, John G.; Holt, Noel R.; Krieg, Peter H.A. Sneath (1994). Bergey's Manual of Determinative Bacteriology, 9th ed. Lippincott Williams & Wilkins. ISBN 0-683-00603-7.
- Madigan, Michael T.; Martinko, John; Parker, Jack (2004). Brock Biology of Microorganisms, 10th Edition. Lippincott Williams & Wilkins. ISBN 0-130-66271-2.
External links
[edit | edit source]- The Infectious Disease Society of America
- GIDEON - GIDEON-Global Infectious Disease Epidemiology Network|Global Infectious Diseases and Epidemiology Network
- When Insects Spread Disease, discusses problem and offers tips for prevention.
- Bacteria of Medical Importance in Todar's Online Textbook of Bacteriology.