Statistics/Analysis of Tuberculosis
Some facts about Tuberculosis
[edit | edit source]Tuberculosis (TB) is a common and deadly infectious disease that undergoes a resurgence nowadays. The WHO estimates that over one-third of world´s population now carries the bacterium in their bodies. People who are HIV-positive face a much higher risk being infected with TB bacilli because their immune systems are compromised by the HIV. Other indicators which may favour the prevalence of TB are bad alimentation, insufficient hygienic circumstances combined with nonexistent medical measures.
Abstract
[edit | edit source]Being neglected for a long time tuberculosis reached a pandemic dimension in many regions around the world. What reasons may there be for such a spread? As mentioned above, there seems to be a consensus about the majority of factors that favour this disease. Nevertheless new hypotheses about the relation between the prevalence of tuberculosis and certain factors have to be quantified by the means of statistical methods, which are carried out with statistical software.
Thus, according to a famous saying from Confucius the goal of our analysis is the way, to be more precise, the way how to assess and deal with multivariate datasets. Nevertheless we will develop some hypotheses about the relations of our variables in the course of the analysis. For a quick glimpse check the section about multivariate analysis and its subsections.
The program used for this analysis is the MDTech XploRe Software. All major steps of the analysis can easily be reproduced by the program code provided in this study (usually together with the graphics in the image space). All you need is the original data called "datest.csv", which is availabale on http://www.quantlet.org/mdbase/, and the free academic version of the software XploRe.
Description of the Original Data Set
[edit | edit source]The analysis is based upon a dataset called "datest.csv", which is available on the homepage of MD*Base.
The missing values that appeared in the original dataset (datorg.csv) have been replaced by estimated values using different methods, like the mean of neighbouring countries, linear regression and other techniques. Since the data was collected from the UN you may have a closer look at the definitions of each variable at the homepage of the United Nations Statistics Division.
The dataset contains 163 observations and 16 variables. The first variable is a text-variable showing the names of the countries, the second variable assigns each country to a continent and is therefore nominal (from one to six). All other variables are described in the following Table:
| No. | Title | Type | Remarks |
|---|---|---|---|
| 1. | Country | Text | Name of each country |
| 2. | Continent | Nominal | 1: Asia, 2: N.America, 3: S. America, 4: Africa, 5: Europe, 6: Australia & Oceanian |
| 3. | Population | Metric | Total population of each country |
| 4. | Condomuse Ratio | Metric | Condomuse in relation to other contraceptives used by women in percent |
| 5. | Aids est. Deaths | Metric | Total number of aids related deaths (estimated) |
| 6. | Malaria | Metric | Prevalence in total numbers |
| 7. | Tuberculosis | Metric | Prevalence per 100.000 people |
| 8. | Drugs | Metric | Access to essential drugs (as listed by the WHO) in percent |
| 9. | Education | Metric | Enrollment Ratio per relevant age group, classified into four groups:
< 50 %, 50 – 80 %, 80 – 95 %, > 95 % |
| 10. | Literacy | Metric | Rate given in percent |
| 11. | Sanitation | Metric | Access to basic sanitation in percent |
| 12. | Water | Metric | Access to improved drinking water in percent |
| 13. | CO2 | Metric | In tons per capita |
| 14. | Internet | Metric | Total number of internet accesses |
| 15. | PC-User | Metric | Total numbers |
| 16. | Telephone | Metric | Total numbers |
Transformation of the Data
[edit | edit source]As you can see from table 1 the variables are measured on a different scale. Tuberculosis is measured per 100.000 people while for example the estimated aids related deaths are given in total numbers of the population. Thus, the first step of our analysis is to rearrange the dataset such that it is more clearly ordered and scaled in a more appropriate way.
The program below is used to:
- add a column with a country code to the dataset
- standardize the scale of all three diseases to "per 100.000"
- change total numbers of internet accesses, personal computers and telephone lines to relative numbers
- rearrange the order of variables such that:
- the first to third column contain countrycode, continentcode and population
- the fourth to sixth column contain standardized values for aids estimated deaths, malaria prevalence and tuberculosis prevalence, respectively
- the variable "condom use" appears in the eighth column, followed by the rest of the variables in original order
The program will load the original data and create two CSV-files that contain the rearranged dataset (including country names) and the country names only. Before you run the program, make sure that you have downloaded the original data "datest.csv" from the homepage of MD*Base to a known directory.
library("xplore")
library("stats")
; ----- Reading data ----------------------------------------------------------------------------
choose = "Read from:" | "Save as:" | "Save country info as:"
defaults1 = "C:\Dokumente und Einstellungen\All Users\Desktop\datest.csv"
defaults2 = "C:\Dokumente und Einstellungen\All Users\Desktop\UN_data_ordered.csv"
defaults3 = "C:\Dokumente und Einstellungen\All Users\Desktop\country.csv"
defaults = defaults1 | defaults2 | defaults3
v = readvalue(choose, defaults)
; ----- Transformation --------------------------------------------------------------------------
x=readcsvm(v[1])
num=1:163
data=num~x.double
country=x.text
pop=data[,3]
x=(data[,5|6]/pop)*100000
y=data[,14|15|16]/pop
data=data[,1:3]~x~data[,7|4|8:13]~y
l=list(country, data)
; ----- Saving ----------------------------------------------------------------------------------
writecsv(l,v[2])
writecsv(country,v[3])
Univariate Analysis
[edit | edit source]To start with we would like to present an overview of some descriptive data on the explanatory variables considering the five number summary, skewness and kurtosis.The skewness and the kurtosis measure the skewness and the departure from the normal distribution, respectively:
;
The skewness should be close to 0 for a distribution that is symmetric around . The kurtosis should be close to 3 for a distribution resembling the normal.
| Variable | Min | 25% Quartile | Median | Mean | 75% Quartile | Max | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|
| Condomuse Ratio | 0 | 4.25 | 6.8 | 10.13 | 12.95 | 77.6 | 2.88 | 18.17 |
| Drugs | 50 | 80 | 80 | 81.47 | 95 | 100 | -0.78 | 2.42 |
| Education | 13.9 | 73.5 | 91.1 | 83.63 | 97.3 | 109.5 | -1.33 | 4.22 |
| Literacy | 24.5 | 83.4 | 95.6 | 88.34 | 99.3 | 100 | -1.64 | 5.12 |
| Sanitation | 8 | 62 | 87 | 76.56 | 98 | 100 | -0.96 | 2.71 |
| Water | 24 | 71.5 | 87 | 81.06 | 98 | 100 | -0.99 | 2.97 |
| Co2 | 0.02 | 0.41 | 1.97 | 4.67 | 6.26 | 90.74 | 6.52 | 61.37 |
| Internet | 0 | 0 | 0.03 | 0.1 | 0.1 | 0.58 | 1.73 | 4.7 |
| PC-User | 0 | 0 | 0.03 | 0.1 | 0.11 | 0.60 | 2.02 | 6.23 |
| Telephone | 0 | 0.03 | 0.22 | 0.4 | 0.55 | 1.61 | 1.16 | 3.01 |
Boxplots, Histograms and Quantile-Quantile Plots
[edit | edit source]With conventional mechanisms of the univariate analysis we will now have a look on all variables of interest for our later model. For an integrated overview, we first focus on multi-graphic displays that contain information about more than just one variable.

Let us have a look at graphic 1 and graphic 2, in which we computed the boxplots and histograms for the diseases in our data namely tuberculosis, malaria and AIDS.
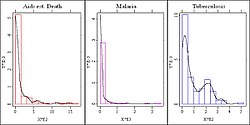
As mentioned above all three variables had been transformed to the scale per 100.000 people. For better visualisation in graphic 1 we standardized the x-axis, otherwise the boxplots for tuberculosis and malaria would have been too compressed. For all three variables we observe skewness to the right nevertheless the characteristic of the outlier distribution differs a lot. So for tuberculosis we only identified one outlier (Cambodia) while for the AIDS-deaths overall 38 outliers are displayed of which 24 are moderate (circles) and 14 are more distinct (crosses) ones. We decided to identify these outliers and it turns out that about 90% of the outliers are African countries and accordingly about 72% of African countries are outliers. Shall a whole continent be excluded here? Definitely not but this fact provides us with a hint for possible subgroups within the data.


Grapics 3 to 5 provide us with the results of our univariate analysis of possible explanatory variables on our dependent variable tuberculosis.
In graphic 3 we see the boxplots for those mentioned. Be aware that we displayed the boxplots on different scales. The upper five boxplots are measured in percent while we decided to standardize the y-axis for the lower boxplots. One important remark has to be done on the boxplot for access to drugs: Although it seems there is only one outlier it turns out that this "point" are altogether 37 countries falling into the 0% - 50% class.
The skewness of the data can also be anticipated and is proved in graphic 4, where we display the average shifted curve together with the histograms. Again what catches the eye is the fact that the upper variables, except for access to drugs, are skewed the left while the lower are skewed to the right. That means the majority of countries have relatively high values for the variables displayed in the upper part and relatively low values for those variables displayed in the lower part. graphic 5 displays the quantile-quantile plots which are used to compare each variable with the normal distribution. Apparently we have a clear deviation from the 45° line which indicates that the variables are not normally distributed.
Altogether, the main finding of the univariate analysis is that we have very skewed distributions, which in part coincide among the different variables. This might be an indicator for strong correlations between the single dimensions. Anyway, the question remains whether these relations are linear or not. Furthermore, we have seen many outliers in the different dimensions of the dataset. Thus, the question arises whether the outliers of one dimension are also outliers in other dimension of our data. This question will be dealt with in the next section.
Outlier Treatment with Simple Multivariate Methods
[edit | edit source]As we have seen in the univariate analysis, we face a very heterogeneous dataset with extremely skewed distributions and therefore with many observations being displayed as outliers. This goes up to the point, where almost a whole continent would be excluded from the analysis, namely Africa in terms of aids related death rates. This might motivate a separate analysis of Africa.
| Population | Aids | Malaria | Tuberculosis | Condom Use | Drugs | Education |
|---|---|---|---|---|---|---|
| 20 | 24 | 37 | 1 | 9 | 31 | 7 |
| Literacy | Sanitation | Water | CO2 | Internet | PC | Telephone |
| 9 | 0 | 3 | 8 | 26 | 18 | 10 |
But here, our goal is to find methods to better handle multidimensional large datasets. Therefore, we try to find a possibility to assess all countries in terms of their extreme values in certain dimensions. Furthermore, we want to get a table with the numbers of "boxplot outliers" in each dimension. Therefore we computed a 163 x 14 matrix containing logical values 0 or 1, where 1 means observation is an outlier. Simple computations using this matrix lead among others to the number of outliers in each dimension shown in table 3.

Graphic 6 shows a barchart, where countries are classified according to the number of univariate extreme values or "boxplot outliers". This charts yields the information that only a few countries are outliers in the boxplot sense in four dimensions, and no country has more than four extreme values. The "outlier program" that generates graphic 6 furthermore provides the option to decide in how many dimensions an observation has to be a univariate outlier to be considered as "multidimensional outlier". These "multidimensional outliers" are then colored blue and shown together with the other observations in a stardiagram (alternativlely one could also choose so called Chernoff-Flurry faces). This facilitates the decision whether those observations are actually really different from the rest of the data. This process can be repeated several times until one finds a satisfying set of outliers that shall be excluded from the further analysis (saving option available via "outlier program").
For the further analysis however, we decided not to exclude any observations, but to proceed with the whole dataset, since even four exceptionally high or low values per observation are still relatively few in comparison to a total of 13 relevant dimensions. Furthermore, the outlier starplot shows that there seem to be different groups of countries in the data that have similar characteristics reflected by the respective shape of the stars. In case we choose to color all observations with one or more "boxplot outliers", the remaining (green) observations seem to have quite similar characteristics. But the number of remaining countries is very limited and does not seem to be a representative group of countries in the world.
Nevertheless, we could check the influence of excluding certain "multidimensional outliers" in the further course of our analysis.
Bivariate Analysis
[edit | edit source]
Now we would like to get a better impression of the relations between our variable of interest, namely the tuberculosis prevalence, and the other variables, which are according to our goal considered to be explanatory variables. One possibility to visualize the relations between all variables within a dataset would be a scatterplot. In such a graphic all variables would be plotted against each other. Since we have 13 variables of interest, this would give us a 13 x 13 display of 2-dimensional plots, which hardly allows for proper displaying on a standard computer monitor. Additionally, we have empty spaces on the diagonal as well as doubling of the same information in the upper and lower triangular. Thus, this graphic can only be used in a proper way for up to eight variables.

Instead, we just plottet all explanatory variables against tuberculosis and display them in one window that is shown in graphic 7. This graphic provides the required informations to derive basic assumptions about the relations within the data.
First, one can see that the majority of observations seem to be distributed in a very small area, in most cases one corner of the plot, whereas only a relatively small proportion is scattered over the whole range of the diagrams. To better visualize this, we added one dimension to plots and computed a two-dimensional density estimate of the plots. This can be seen in graphic 8, which exemplary shows such a two-dimensional density estimate for "tuberculosis" and "sanitation". This further strengthens the idea mentioned in the preceding steps of our analysis. That is, the countries can be classified into different groups according to the information/variables available. Furthermore, there seem to be different relations between the explanatory variables and tuberculosis. These relationships are to be considered in the following steps of our analysis. Since they may differ from subgroup to subgroup, we proceed with trying to find homogeneous groups within the countries and turn to developing hypotheses on the relationships in the section on multivariate analysis.
Finding Groups
[edit | edit source]Since we have seen many indicators for the existence of different groups, we will now try to find and interpret the groups that can be found in the data. Before we use the statistical methods at hand, we would like to mention a common distinction for the countries around the globe. This is the separation of countries according to their "level of overall development" into developed countries, often used synonymously for western European countries, North America and Japan, emerging countries, like the southeast Asian tiger states, and most of the Latin American countries, and into developing countries, formerly often described as the third world (although there is just one), since these are the poorest countries in terms of income and living standard. Although the distinction between those three groups of countries is not really based upon data like ours, but in fact incorporates much more economical data as well as data from social sciences, one would nevertheless expect to find groups similar to the ones described above. With the statistical method of Cluster analysis, we will now try to find groups that differ from each other as clearly as possible.
Cluster Analysis
[edit | edit source]
The aim of a cluster analysis is to construct groups with homogeneous properties out of a heterogeneous large dataset. The methods used are usually divided into two steps: The choice of a proximity measure which checks each pair of observations (objects) for the similarity of their values. A similarity (proximity) measure is then defined to measure the closeness of the objects. The closer they are, the more homogeneous they are. And the choice of a group-building algorithm which on the basis of the proximity measures the objects assigned to groups so that differences between groups become large and observations in a group become as close as possible.
For our analysis we chose to apply the euclidean distance for our proximity measure which the squared distance between two points. But before doing so we standardized the data by dividing it through the variance because of the different scaling used for the variables.
Our algorithm is the so called Ward clustering algorithm which joins groups that do not increase a given measure of heterogeneity too much. The aim of the Ward procedure is to unify groups such that the variation inside these groups does not increase too drastically: the resulting groups are as homogeneous as possible. We see the graphical representation of the sequence of clustering within the dendogram in graphic 9 It displays the observations, the sequence of clusters and the distances between the clusters. The vertical axis displays the indices of the points, whereas the horizontal axis gives the distance between the clusters.
We can clearly distinguish between three groups (clusters) with relatively high homogeneity. The biggest cluster containing 108 observations on the right is representing the developing countries, the group in the middle with 37 observations stands for the emerging countries whereas the smallest group with overall 18 observations is identical to the developed countries.
Additionally we computed the PCP (Parallel Coordinate Plot) for the three cluster means to visualize the differences between those groups. The green line represents the developed countries, the red line stands for the emerging and the blue for developing countries. As we expected two clearly opposed groups can be observed. The red line (emerging countries) approaches the blue one (developed countries), which seems to be a plausible result.
To get a list of the countries included in the different clusters, run the program that you find attached to graphic 9.
Multivariate Analysis
[edit | edit source]As mentioned in the section on bivariate analysis, different relations between Tuberculosis and the explanatory variables contained in our dataset might be supposed. It should as well be mentioned that we do neither have very sophisticated medical knowledge nor do we know the circumstances of high tuberculosis prevalence countries from our own experience. Thus, it could be difficult to understand the relations between the different variables. Nevertheless we will make some assumptions for almost every explanatory variable and see whether they hold or not. From graphic 5 of the section on bivariate analysis, we found the regression lines (red, solid) shown in graphic 11 to be quite good approximations to the pointclouds. These relations might be explained in the following, possibly naive way.
Hypotheses Development
[edit | edit source]
- Tuberculosis prevalence vs. aids related deaths
- The functional form used here (graphic 10 and graphic 11), is based on the assumption of exponentially increasing numbers of aids related deaths if tuberculosis prevalence rises. Although, both variables are mutually related, medical scientists would probably argue, that aids related deaths are influenced by tuberculosis prevalence rather than the other way around. However, one could (politically probably not entirely correct) argue, that numbers of people infected with tubercolsis decline if more people suffering from both aids and tuberculosis die. Therefore, we assume that this variable can contribute to the explanation of tuberculosis prevelance.
- Tuberculosis prevalence vs. malaria prevalence
- There seems to be no linear relation between malaria and tuberculosis. But if we excluded some of the countries with extremely high cases of malaria, we would probably learn much more. This can easily be done with the "paf" command. Still, there is no significant influence of malaria prevalence on tuberculosis prevalence as you can see from the p-value in the following table.
Contents of out [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 22178.097 1 22178.097 2.228 0.1375" [ 5,] "Residuals 1602701.069 161 9954.665" [ 6,] "Total Variation 1624879.166 162 10030.118" [ 7,] "" [ 8,] "Multiple R = 0.11683" [ 9,] "R^2 = 0.01365" [10,] "Adjusted R^2 = 0.00752" [11,] "Standard Error = 99.77307" [12,] "" [13,] "" [14,] "PARAMETERS Beta SE StandB t-test P-value" [15,] "________________________________________________________________________" [16,] "b[ 0,]= 95.8129 8.1290 0.0000 11.787 0.0000" [17,] "b[ 1,]= 0.0296 0.0198 0.1168 1.493 0.1375"
- Tuberculosis prevalence vs. condom use to contraceptive by women
- As already mentioned in our univariate analysis this variable is difficult to handle, since the relative usage of condoms does not seem to say anything about the frequency of condom use during sexual intercourse. If other contraceptives are not used frequently, the relative condom use ratio might be high. Thus, we did not make assumptions about the relation between these two variables. Nevertheless, it turns out that the coefficient of the condom use ratio in a simple linear regression model on tuberculosis prevalence is significantly different from zero.
Note: If we refer to significance in the further analysis, we always talk about a minimum alpha of 5%! For space reasons we abstain from integrating all regression output tables.

- Tuberculosis prevalence vs. access to essentiell drugs
- The difficulty here is that we have classified data. Nevertheless we assume a linear relationship, which is confirmed by a significant regression coefficient. The negative correlation is obvious.
- Tuberculosis prevalence vs. education ratio and literacy rates
- Although we will probably have heteroscedasticity, we assume a linear relationship for the whole dataset. This is also confirmed by significant regression coefficients.
- Tuberculosis prevalence vs. access to sanitation and clean drinking water
- Again heteroscedasticity has to be taken into account. The assumption about a linear relationship is confirmed by a significant p-value.
- Tuberculosis prevalence vs. CO2 emissions
- A plausible explanation for a relationship between CO2 emissions and tuberculosis prevalence would be a rather difficult construct. But if we consider CO2 emissions as latent factor for the general economic development and living standard, the relationship seems to make more sense. The functional form assumed and displayed in graphic 11 (3rd row, first element) is justified by the assumption of negative marginal influence, i.e. for a low level of CO2/living standard the influence of a little increase in the CO2 emissions/living standard on tuberculosis prevalence is stronger and diminishes from a certain level onwards.
- Tuberculosis prevalence vs. internet accesses, PCs, and telephones p.c.
- These three variables could be interpreted as access to information. We assume that given a low level of overall information additional information is especially valuable, take for example the information that you can get infected with tuberculosis via human fluids, a very basic information that can easily be broadcasted via the media. Therefore, information certainly has a stronger influence on tuberculosis prevalence if the overall level of available information is low. If people are already well supplied with news, additional information might just cause what we would call information overflow, i.e. they are not realized anymore.
- The functional form used to describe the relationship between these four variables is, for simplicity reasons, the same for all of them. The justification basically is the assumption of negative marginal influence for all variables as explained above. The functional form used here is:
- The p-values of the simple linear regression models for the last four (transformed) variables are also significant. And the adjusted R^2 is throughout relatively high as you can see in the output tables below. That is why we will maintain our hypotheses.
Contents of out [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 781723.888 1781723.888 149.270 0.0000" [ 5,] "Residuals 843155.278 161 5236.989" [ 6,] "Total Variation 1624879.166 162 10030.118" [ 7,] "" [ 8,] "Multiple R = 0.69361" [ 9,] "R^2 = 0.48110" [10,] "Adjusted R^2 = 0.47787" [11,] "Standard Error = 72.36705" [12,] "" [13,] "" [14,] "PARAMETERS Beta SE StandB t-test P-value" [15,] "________________________________________________________________________" [16,] "b[ 0,]= -21.0144 11.3520 0.0000 -1.851 0.0660" [17,] "b[ 1,]= 118.3640 9.6880 0.6936 12.218 0.0000" Contents of out [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 605005.143 1605005.143 95.508 0.0000" [ 5,] "Residuals 1019874.023 161 6334.621" [ 6,] "Total Variation 1624879.166 162 10030.118" [ 7,] "" [ 8,] "Multiple R = 0.61020" [ 9,] "R^2 = 0.37234" [10,] "Adjusted R^2 = 0.36844" [11,] "Standard Error = 79.59033" [12,] "" [13,] "" [14,] "PARAMETERS Beta SE StandB t-test P-value" [15,] "________________________________________________________________________" [16,] "b[ 0,]= 6.5870 11.3392 0.0000 0.581 0.5621" [17,] "b[ 1,]= 25.3361 2.5925 0.6102 9.773 0.0000" Contents of out [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 663303.291 1663303.291 111.059 0.0000" [ 5,] "Residuals 961575.874 161 5972.521" [ 6,] "Total Variation 1624879.166 162 10030.118" [ 7,] "" [ 8,] "Multiple R = 0.63892" [ 9,] "R^2 = 0.40822" [10,] "Adjusted R^2 = 0.40454" [11,] "Standard Error = 77.28209" [12,] "" [13,] "" [14,] "PARAMETERS Beta SE StandB t-test P-value" [15,] "________________________________________________________________________" [16,] "b[ 0,]= -18.9252 12.7351 0.0000 -1.486 0.1392" [17,] "b[ 1,]= 35.7036 3.3879 0.6389 10.538 0.0000" Contents of out [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 805435.082 1805435.082 158.248 0.0000" [ 5,] "Residuals 819444.083 161 5089.715" [ 6,] "Total Variation 1624879.166 162 10030.118" [ 7,] "" [ 8,] "Multiple R = 0.70405" [ 9,] "R^2 = 0.49569" [10,] "Adjusted R^2 = 0.49256" [11,] "Standard Error = 71.34224" [12,] "" [13,] "" [14,] "PARAMETERS Beta SE StandB t-test P-value" [15,] "________________________________________________________________________" [16,] "b[ 0,]= -25.7826 11.3957 0.0000 -2.262 0.0250" [17,] "b[ 1,]= 61.4014 4.8810 0.7041 12.580 0.0000"
Multiple Linear Regression Models
[edit | edit source]Now, what happens if we try to put all explanatory variables into one model? Will their influence still be significant in relation to the impact of other variables? We will try different selection processes implemented in XploRe to compute the model with the best fit, i.e. with the best adjusted .
Forward Selection Model
[edit | edit source]The forward selection option starts from one "good" variable, calculates the simple linear regression and then decides stepwise for each variable if its inclusion can improve the fit of the model.
This process yields the following model as result:
Contents of out [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 910056.048 2455028.024 102.567 0.0000" [ 5,] "Residuals 700949.728 158 4436.391" [ 6,] "Total Variation 1611005.776 160 10068.786" [ 7,] "" [ 8,] "Multiple R = 0.75160" [ 9,] "R^2 = 0.56490" [10,] "Adjusted R^2 = 0.55939" [11,] "Standard Error = 66.60624" [12,] "" [13,] "" [14,] "PARAMETERS Beta SE StandB t-test P-value" [15,] "________________________________________________________________________" [16,] "b[ 0,]= -29.2719 10.7465 0.0000 -2.724 0.0072" [17,] "b[ 1,]= 12.7427 2.5019 0.3098 5.093 0.0000" [18,] "b[ 2,]= 47.5180 5.2959 0.5458 8.973 0.0000"
Backward Elimination Model and Stepwise Selection Model
[edit | edit source]The backward elimination process starts from the full multiple regression model and stepwise excludes variables that do not contribute much to the model fit. The stepwise selection model in our case yields the same results as the backward elimination. Both lead to the following model:
Contents of ANOVA [ 1,] "" [ 2,] "A N O V A SS df MSS F-test P-value" [ 3,] "_________________________________________________________________________" [ 4,] "Regression 963289.397 4240822.349 61.982 0.0000" [ 5,] "Residuals 606118.006 2e+02 3885.372" [ 6,] "Total Variation 1611006 160 10068.786" [ 7,] "" [ 8,] "Multiple R = 0.77327" [ 9,] "R^2 = 0.59794" [10,] "Adjusted R^2 = 0.61412" [11,] "Standard Error = 62.33275" Contents of Summary [ 1,] "Variables in the Equation for Y:" [ 2,] " " [ 3,] "" [ 4,] "PARAMETERS Beta SE StandB t-test P-value Variable" [ 5,] " __________________________________________________________________________________" [ 6,] "b[ 0,]= 155.7053 41.5609 0.0000 3.7464 0.0003 Constant " [ 7,] "b[ 1,]= 10.5762 2.3854 0.2571 4.4337 0.0000 X 1" [ 8,] "b[ 2,]= -0.8998 0.3050 -0.2283 -2.9500 0.0037 X 7" [ 9,] "b[ 3,]= -0.8339 0.4242 -0.1621 -1.9659 0.0511 X 8" [10,] "b[ 4,]= 26.3803 6.6151 0.3030 3.9879 0.0001 X 12"
This is not a very satisfying result, since the explanatory power of the model is not substantially higher than most simple regression models. What are the reasons for the lack of fit? This question shall be addressed in our final conclusion.
Conclusions
[edit | edit source]As we have seen in the course of our analysis, there are many possibilities to deepen your understanding of an unknown dataset. Although we did not really find a satisfying model that fully explains the differences in tuberculosis prevalence across the countries, we got a much better idea of the structures within the dataset. We have seen and tried to explain single bivariate relations between the tuberculosis prevalence and all other variables. Furthermore, we found quite reasonable groups in the data that could be assessed seperatly via the programs provided.
However, a multivariate analysis like a multiple regression requires more sophisticated methods. As we can easily see by looking at the correlation matrix of the whole dataset, many of the different variables are correlated with each other. This is obvious in some cases, e.g. in terms of telephone lines, internet access possibilites and personal computers. These multicollinearity issues make the multiple regression rather difficult, since numerous models with similar fits but different explanatory variables can be obtained. Thus, we should try to find a possibility to reduce the dimensions of the dataset, e.g. with a factor analysis.
Finally, we should mention again that our hypotheses are neither based on sophisticated medical expertise nor can we say anything about the developement of our data in time, which might be very helpful to derive better assumptions that could be checked with the methods presented.
Anyway, we left the reader himself with several opportunities to continue the analysis with the help of the programs provided. One could for example, use the different clusters (save them via program 9) to repeat the outlier treatment (with program 6) or the bivariate analysis (with program 11), etc.
References
[edit | edit source][Härdle, Klinke, Müller 2000] Härdle, W.; Klinke, S.; Müller, M.: Xplore Learning Guide. Springer Verlag Berlin-Heidelberg, 2000
[Härdle, Simar 2003] Härdle, W.; Simar, L.: Applied Multivariate Statistical Analysis. Springer Verlag Berlin-Heidelberg, 2003
[Hädle, Hlavka, Klinke 2000] Hädle, W.; Hlavka, Z.; Klinke, S.: XploRe Application Guide. Springer Verlag Berlin-Heidelberg, 2000
United Nations Statistics Division, unter http://unstats.un.org/unsd/cdb/cdb_list_dicts.asp , zugegriffen am 9.12.2006
XploRe Help, unter http://www.xplore-stat.de/help/_Xpl_Start.html


![{\displaystyle s=\left[{\frac {1}{n}}\sum _{i=1}^{n}{\frac {(x_{i}-{\bar {x}})^{3}}{v^{3}}}\right]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/37a95e66a8853c3551d9ad228b7afd81a4a53ee1)
![{\displaystyle k=\left[{\frac {1}{n}}\sum _{i=1}^{n}{\frac {(x_{i}-{\bar {x}})^{4}}{v^{4}}}\right]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/e87e6f2ac900bd93f8a946b78ef3e4f2b3025f02)




