Sensory Systems/Visual System/In Vivo Imaging
Allen Institute Big Data
[edit | edit source]There are approximately 100 billion neurons in the brain. The complexity within the brain is unparalleled. To give an example, when changing images are displayed to passive mice, there are excitatory neurons, inhibitory neurons, neurons that increase activity when predicted image change occurs, and neurons that build up activity continuously when a predicted image change does not occur. With this dimensionality of information encoding schemes, which must be integrated downstream to form our complete visual perception, a key goal within the neuroscience community has been the production of big data. To succeed at producing in vivo data of sensory processing on a very large scale, institutions such as the Allen Institute for Brain Science have spent the past decade focalizing pipelines that automate data production and collection. As of today, the Allen Institute has had numerous massive releases of data generated from the Visual Coding pipeline and is currently undertaking a new approach toward data production through their Visual Behavior pipeline.
The Allen Institute is a key example of implementing successful imaging strategies to generate massive quantities of sensory processing data through standardized procedures. There are seven steps to the Allen Institute’s pipeline, and teams of engineers, optical technicians, and surgical technologists who collaborate to keep it functioning smoothly. This integration of disciplines and minds has as of yet produced morphology, gene activity, electrophysiology, calcium imaging, and topology data on the order of 1900 cell types. All data is collected using cutting edge imaging techniques, which make long term and highly sensitive recordings possible.
Experimental Setup
[edit | edit source]Achieving high resolution throughout any in vivo imaging process requires a few unique heuristics. Before any imaging can occur, the mouse has to undergo a surgery to implant a headframe. During the procedure, surgeons remove a small section of skull, which is then covered with a glass cranial window attached to the headframe (Figure 1). The headframe’s purpose is twofold: (1) it permits visibility of fluorescence changes during the imaging step, and (2) it may be hooked onto a head-plate, ensuring head fixation and highest resolution possible during the imaging. Since this fully constrains the head and increases chances of stress, a rotating disk is positioned under the mice to allow stress-relieving running activity (Figure 2). This activity is monitored and entered in the data output, as an additional analytical technique for researchers. Other data measures recorded include videographic assessment of pupil dilation, body position, running data, and eye tracking, indicated in post-assessment as a red cross on the image screen corresponding to the point of gaze of the mouse[1]. Past experimental data has revealed that mice generally have far less eye movement when exposed to a new image than humans, since their foveal vision has approximately the same resolution as their peripheral vision.
Visual Coding pipeline
[edit | edit source]The Allen Brain Observatory Visual Coding pipeline takes place in (7) steps, delineated below.
- Cre driver x GCaMP6 reporter: The transgenic mouse lines are genetically engineered to express the GCaMP6 genetically encoded calcium indicator (GECI)[1]. GECIs have localized expression depending on the specified cell lines or functional areas in which they are encoded. Thus Allen geneticists effectively highlight the brain for further imaging steps by selecting genes, editing them by inserting GCaMP6, and driving expression of the gene.
- Surgery: Mice undergo a surgical procedure for headframe implantation. If the stability of the headframe is deemed unsuitable for two photon calcium imaging, a second surgery is prescribed in order to apply additional Metabond to the headframe.
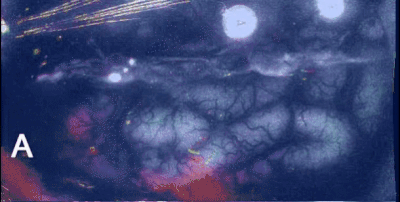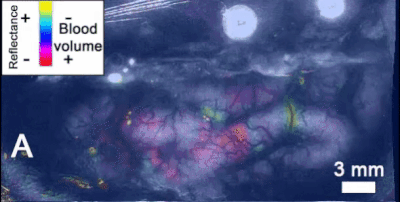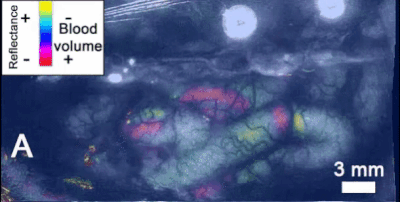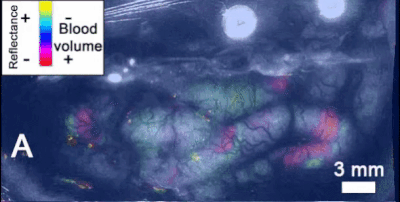
- Intrinsic signal imaging (ISI): ISI is a tool to measure hemodynamic response, i.e., the reflectance of hemoglobin that is directly related to oxygenation levels. Allen scientists use ISI in this step to map functionally defined cortical areas and retinotopic organization. Mice are exposed to visual stimuli of black and white checkerboards, translating in all directions along the cardinal axes, on the scale of the entire visual field. Then, phase maps are calculated of the visual stimuli using a discrete fourier transform parametrized by the input stimulus frequency. Phase information is useful for mapping visual stimuli into cortical areas such as V1. Azimuth and altitude maps are also procured by projecting along the azimuth/altitude respectively, so information about the azimuth/altitude distances with respect to a central point are conserved. Sign maps are generated by taking the sine of the azimuth and altitude gradients of the phase map. In order to standardize this technique, each new mouse is compared to a previously defined generalized model of cortical visual areas, based off of 35 combined sign maps (Figure 4). An automated process optimizes the sign map fit of the new map to the old model using size, phase, location, and spatial relationship information of each defined visual area[1]. Vasculature is also used to determine correct placement of visual cortex boundaries. This results in a common coordinate framework (CCF) mapping, which is highly useful in the following step of two-photon imaging.
- Habituation: The mouse is habituated to the experimental set up over a ten-day period, with passive viewing of images to reduce stress and allow for acclimation. Throughout this time period the mouse is exposed to the full range of stimuli images. Stress is monitored via nose bulge, excessive secretion around the eye, and/or abnormal posture[1].
- In vivo two photon calcium imaging: Hardware includes a Nikon A[1]R MP+ two-photon imaging microscope and a laser at 910 nm. Deconvolution calculations are applied to the data following imaging, due to fluorescence leakage between pixels, i.e. fluorescence being carried from the first imaged pixel to the next[1]. The convolutional equation is characterized below:
Where the symbols denote:
This equation accounts for non-uniformity of leakage, given empirical findings of lesser leakage around the edges and greater leakage at the center.
Laser power is optimized between two constraints, maximum power and avoidance of pixel saturation. Over an experimental session period of ~90 minutes, a z-stack of images of approximately .1 micrometers is produced.
Region of interest (ROI) filtering was also performed, using a variety of filters to create a binarized mask of active cells vs. background noise. No data reduction techniques are implemented, rather, band pass filtering is first applied, and then the full image is subtracted from itself passed through a low pass filter (Figure 3). Further analysis included classification analysis, using parameters including object shape, size, and area as found in previous data set samples. Once ROI’s are derived, the image can be segmented into its cell objects and background, and only fluorescence from these within the cell objects is passed forward to be summed by the photomultiplier tube (PMT).
- Intrinsic Signal Imaging: Verification that the functionally defined visual cortex areas remain centered on the CCF.
- Serial Two-Photon Tomatography: Tissue imaging of the mice brain to infer the 3D volume of brain images for all cortical regions and detect structural heterogeneity.
|
 |
All information on the Visual Coding Pipeline is explained in detail in the Allen Institute white papers.
Calcium Indicators
[edit | edit source]
Calcium dynamics are tightly coupled to action potentials conducting through neurons. The action potential, defined broadly as a rapid membrane depolarization and consequent hyperpolarization sequence of events, are generated by ions and ion channels. Voltage gated ion channels are the chauffeurs of sodium and potassium cations, and open with increasing membrane voltage potential via a positive feedback mechanism. The sequence of events for an ion potential then, is generally immutable: after a certain influx of sodium ions surpasses a threshold, all of the channels will eventually open, and the membrane potential will reach a maximum correlating to the cell firing. The sodium voltage gated channels conclude the action potential by closing simultaneously with the slower potassium channels open, so potassium ions may flow out and restore the resting state electrochemical gradient. However, this electrical communication is confined to the membrane of a single neuron, since neurons are not physically connected, and thus gap junctions between neurons act as electrical shorts. Chemical synaptic transmission is required to carry the signal between the gap junctions. Neurotransmitters fill this role and are intrinsically linked to calcium dynamics. In a resting state neuron, neurotransmitters are held within vesicles at the presynaptic active zone. When an action potential is fired, it opens calcium ion channels, and incoming calcium ions flow into the neuron as the action potential proceeds(Figure 5). These ions trigger migration of neurotransmitter-filled vesicles toward the cell’s surface, where they release their contents[2]. Thus, calcium ions are a proxy of action potentials, and monitoring their dynamics provides fundamental insight into neuronal activity. It is also worth noting that calcium ions are a salient factor in neural plasticity and neural development[3] in the beginning stages of a neuron’s life.
Calcium indicators have a multifaceted purpose – to link calcium to fluorescence, and to constrain this linkage to a localized area. There are two fields of calcium indicators: chemical indicators and genetically encoded calcium indicators (GECI)[4]. Chemical indicators chelate to calcium ions and are typically introduced to specific neuronal populations through a loaded calcium dye[5]. There are several drawbacks with this strategy: (1) chronic imaging of the same neural populations requires repeated injections of calcium dye and is prone to human error, and (2) specificity of recordings are limited to spatial locations, rather than cell types or subcellular compartments[6]. GECIs maneuver around these shortcomings while detecting calcium dynamics at comparative sensitivity levels. Since they are genetically encoded, they may be used in chronic imaging studies, which has enormous benefits. Producing long term, consistent information originating from a single brain region in one mouse is essential in producing big data. There are an additional two classes of GECIs in existence: (1) Cameleon proteins, and (2) GFP fluorophores. Cameleons undergo a conformational change upon calcium binding and radiate at a different wavelength. GFP fluorophores, on the other hand, feature a Ca2+ dependent protein encoded as a sequence insert into the GFP fluorophore genetic code[7]. The GFP fluorophore GCaMP6 is used in the Allen Visual Coding pipeline.
Imaging Microscopy Techniques
[edit | edit source]After the reporter molecules for fluorescence expression have been chosen, the imaging technique must be decided upon. There are two major camps of calcium fluorescence imaging: (1) Fluorescence microscopy, and (2) two photon calcium imaging. The prior, fluorescence imaging, utilizes light scattering effects. It can be used to image a thin section of fixed tissue and records the emitted light of a surface illuminated by a laser with a charge coupled device (CCD) array. However, it lacks spatial specificity and depth due to this light scattering based measurement, which is non-trivially affected by signal noise from proteins and other small molecules at depths of greater than 100 microns. Two photon calcium imaging is useful as it surpasses these limits, achieving depths of greater than 100 microns and enhancing spatial specificity (Figure 6).
The discrepancy between two photon calcium imaging and epifluorescence is engineered via the fluorophores: in two photon calcium imaging, fluorophores are protein engineered to require twice the wavelength to be excited (~1240 nm vs. ~620 nm), raising their excitation range to the infrared spectrum. Thus, excitation is achieved by two coincident photons arriving at a location, i.e. at the intersection of two lasers, with resolution on the order of a micron. Imaging information is then obtained per micron step by measuring the emitted photons with a photomultiplier tube (PMT). This photon signal is converted into a voltage, summed over the time period of data collection, and assigned to a pixel corresponding to that location in the final image.
Imaging vs. Electrophysiology
[edit | edit source]Imaging is a relatively new technique compared to electrophysiology, and contrasts in a number of ways due to the different nature of the information recorded. Imaging recording has a different spatial focus, as it generates x-y dimensional information at a z depth, while most electrode related electrophysiological techniques generate z dimensional depth information. This must be weighed by the researcher in context with his experimental objective. There are also intrinsic drawbacks with each technique unlikely to be completely improved. Imaging recording is almost always an indirect measurement, as in the calcium imaging method detailed above. While calcium is a consequence of a traveling action potential, it is not the driver behind it. There is also inability to measure sub-threshold voltages with calcium imaging, since it is limited to the triggering of calcium influx following an action potential. However, calcium imaging is generally less mechanistically intrusive, has more breadth, and can generate in vivo data of z stacked layers of neural activity. This allows for simultaneous imaging and behavioral data collection, discussed below. It also takes biochemical information of calcium indicator locations into account, unlike electrophysiological recordings, which are constrained to electrical signaling information of spiking activity or compound field potentials.
One important consideration in every experiment is signal-to-noise ratio, which has different sources in imaging vs. electrophysiological recordings. The difference is in the transduction of information required in imaging, where electrical or chemical signals are translated into photons measured by the difference in fluorescence over the original fluorescence. Electrophysiological recordings, such as the patch clamp technique, are inhibited by instrumental cofactors. On the other hand, the noise perpetrators in imaging are the discrete photons that cause shot noise when relatively low levels are being detected, resulting in large fluctuations around the mean signal produced[7]. This shot noise can be reduced by increasing the concentration of reporter molecules expressing fluorescence, so long as the biological system is not perturbed.
Visual Behavior pipeline
[edit | edit source]Sensory processing is inherently linked to awareness, since sensory processing may be modulated by top down influences such as brain states. For example, different neurons fire in response to structurally similar images depending on awareness and prediction of what is occurring in the images. A recognizable object in the field of view will set off a cascade of action potentials down the ventral processing stream. Expectation of visual stimuli that is unfulfilled results in increased calcium influx, i.e. firing, in certain neurons. Observation of a small animal from far away while commuting to work in Switzerland’s temperate climate will generally be detected as a cat or squirrel rather than a koala.
Neurons integrate feedforward and feedback information into their properties[8], and thus we cannot fully understand neural activity in visual encoding without attempting to understand broader contextual influences such as awareness. In lieu of this information, providing some means for the experimental animal to communicate awareness is highly salient. This communication, easy enough with human participants, requires behavioral training in mice and other animals.
The Allen Institute has implemented behavioral analysis via the Visual Behavior pipeline. The Visual Behavior pipeline is conducted through the same sequence of steps as the Visual Coding pipeline, with the added step of behavioral training following habituation.
Behavioral training of image detection occurs in four steps, with the end goal of teaching the mouse to respond to an image change with operant conditioning. Operant conditioning incorporates delivery of a reward if the correct behavior is demonstrated. The lickspout is introduced in the free rewards phase following habituation. The third phase is static gratings visual stimuli and contingent rewards phase. Thus, in this stage the task at hand is identification of static grating rotation (from 0 degrees to 90 degrees) and generally results in high accuracy once the mice are properly trained. At the initial onset of this stage, excessive or exploratory licking is usually observed, and is an indicator that the mouse is not properly trained as of yet.
Depending on the combination of image detection trial and observed behavioral response, there are four types of trials which may be observed. The Hit trial is a combination of a go trial with the trained lick response, which is the desired outcome and is rewarded in training. The Correct Rejection trials are also desired outcomes, and occur when the mouse does not lick for a catch trial. There are two types of incorrect responses, however: miss trials – “go” trial, no lick – and false alarm trials – “catch” trial, lick.
With this experimental training set up, the mice learn to respond in image detection tasks, allowing data analysts to correlate neural activity, image sequence, and higher order encoding representation measured via behavioral response. The architecture of the experiment, however, yields several potential pitfalls, which the Allen Institute has controlled for in the past. First, the position of the lick port is surprisingly relevant. The position of the lick port is currently placed using a coordinate system adjusted and then recorded for each mouse. This placement was realized to be important when patterns were observed correlating lick response to lick port placement. When the lick port is too close or too far, the mouse ostensibly weighs a cost function of water payoff with effort to lick. Thus, excessive licking was observed when the lick spout was too close, and hindered licking, when too far. Another relevant feature of this experimental set up shows in the algorithm behind image presentation. The image change cannot be produced from a timed image train or the mouse learns to predict it. Thus, an element of stochasticity must be introduced to keep the mouse – and its neurons – from predicting image change.
References
[edit | edit source]- ↑ a b c d e Allen Brain Observatory. (2017). Visual Coding Overview. https://commons.wikimedia.org/wiki/Commons:Email_templates
- ↑ Südhof, T. C. (2012). Calcium Control of Neurotransmitter Release. Cold Spring Harbor Perspectives in Biology, 4(1): a011353.
- ↑ Yang, S.N., Tang, Y.G., Zucker, R. (1999). Selective Induction of LTP and LTD by Postsynaptic [Ca2+]i Elevation. American Physiological Association, 81(2): 781-787.
- ↑ Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, et al. (2004) Functional Fluorescent Ca2+ Indicator Proteins in Transgenic Mice under TET Control. PLoS Biol, 2(6): e163.
- ↑ Wikipedia contributors. (2018, July 12). Calcium imaging. In Wikipedia, The Free Encyclopedia.
- ↑ Feng et al. (2012). Imaging Neural Activity Using Thy1-GCaMP Transgenic Mice. Neuron, 76(2): 297-308.
- ↑ a b Hausser, M., Scanziani, M. (2009). Electrophysiology in the age of light. Nature. 461(7266): 930-9.
- ↑ Gilbert, C., Sigman, M. (2007). Neuron, 54(5): 677-696.

![{\displaystyle {\begin{array}{lcl}H&=&{\text{leakage filter}}={\text{pixel series (ym)/ time series (yT)}}\\{\tilde {H}}&=&{\text{Fourier transform of H}}\\{\tilde {y}}&=&{\text{pixel series * }}{\tilde {H}}{\text{ + noise}}\\\lambda &=&.[1]={\text{smoothness weight, empirically tested}}\\\Lambda &=&{\text{Laplacian Operator}}\end{array}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/61a21c8dbcfe6ec2f908ab22608bfec69e5236c3)