Principles of Biochemistry/Nucleic acid III: Sythesis of nucleotides
Purines Synthesis
[edit | edit source]Purines are biologically synthesized as nucleotides (bases attached to ribose 5-phosphate). A key regulatory step is the production of ribose-5-phospho-α-D-ribosyl 1-pyrophosphate (PRPP) by PRPP synthetase, which is activated by inorganic phosphate and inactivated by purine ribonucleotides. It is not the committed step to purine synthesis because PRPP is also used in pyrimidine synthesis and salvage pathways. The first committed step is the reaction of PRPP, glutamine and water to 5'-phosphoribosylamine, glutamine, and pyrophosphate - catalyzed by pyrophosphate amidotransferase, which is activated by PRPP and inhibited by AMP, GMP and IMP.
Both adenine and guanine are derived from the nucleotide inosine monophosphate (IMP), which is synthesised on a pre-existing ribose-phosphate through a complex pathway using atoms from the amino acids glycine, glutamine, and aspartic acid, as well as formate ions transferred from the coenzyme tetrahydrofolate[1].
GMP
[edit | edit source]- IMP dehydrogenase converts IMP into XMP
- GMP synthase converts XMP into GMP
In enzymology, a GMP synthase (glutamine-hydrolysing) (EC 6.3.5.2) is an enzyme that catalyzes the chemical reaction
- ATP + xanthosine 5'-phosphate + L-glutamine + H2O AMP + diphosphate + GMP + L-glutamate
The 4 substrates of this enzyme are ATP, xanthosine 5'-phosphate, L-glutamine, and H2O, whereas its 4 products are AMP, diphosphate, GMP, and L-glutamate. This enzyme belongs to the family of ligases, specifically those forming carbon-nitrogen bonds carbon-nitrogen ligases with glutamine as amido-N-donor. The systematic name of this enzyme class is xanthosine-5'-phosphate:L-glutamine amido-ligase (AMP-forming). Other names in common use include GMP synthetase (glutamine-hydrolysing), guanylate synthetase (glutamine-hydrolyzing), guanosine monophosphate synthetase (glutamine-hydrolyzing), xanthosine 5'-phosphate amidotransferase, and guanosine 5'-monophosphate synthetase. This enzyme participates in purine metabolism and glutamate metabolism. At least one compound, Psicofuranin is known to inhibit this enzyme[2][3].
- GMP reductase converts GMP back into IMP
AMP
[edit | edit source]- adenylosuccinate synthase converts IMP to adenylosuccinate
- adenylosuccinate lyase converts adenylosuccinate into AMP
- AMP deaminase converts AMP back into IMP
Phosphoribosyl pyrophosphate (PRPP) is a pentosephosphate.
It is formed from ribose 5-phosphate by the enzyme ribose-phosphate diphosphokinase.
It plays a role in transferring phospho-ribose groups in several reactions:
| Enzyme | Reactant | Product |
|---|---|---|
| adenine phosphoribosyltransferase | adenine | AMP |
| hypoxanthine-guanine phosphoribosyltransferase | guanine | GMP |
| hypoxanthine-guanine phosphoribosyltransferase | hypoxanthine | IMP |
| orotate phosphoribosyltransferase | orotate | OMP |
| uracil phosphoribosyltransferase | uracil | UMP |
In de novo generation of purines, the enzyme amidophosphoribosyltransferase acts upon PRPP to create phosphoribosylamine. Six enzymes take part in IMP synthesis. Three of them are multifunctional:
GART (reactions 2, 3, and 5)
PAICS (reactions 6, and 7)
ATIC (reactions 9, and 10)
Trifunctional purine biosynthetic protein adenosine-3 is an enzyme that in humans is encoded by the gene GART . This protein is a trifunctional polypeptide. It has phosphoribosylglycinamide formyltransferase (EC 6.3.4.13), phosphoribosylglycinamide synthetase (EC 6.3.3.1), phosphoribosylaminoimidazole synthetase (EC 2.1.2.2) activity which is required for de novo purine biosynthesis.
Phosphoribosylaminoimidazole carboxylase (or AIR carboxylase) is an enzyme involved in nucleotide synthesis. It catalyzes the conversion of 5'-phosphoribosyl-5-aminoimidazole ("AIR") into 5-phosphoribosyl-4-carboxy-5-aminoimidazole ("CAIR").
Bifunctional purine biosynthesis protein PURH is a protein that in humans is encoded by the ATIC gene. ATIC encodes an enzyme which generates inosine monophosphate from aminoimidazole carboxamide ribonucleotide. It has two functions: EC 2.1.2.3 - 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase EC 3.5.4.10 - IMP cyclohydrolase
Inosinate synthesis
[edit | edit source]| The biosynthetic origins of purine ring atoms N1 arises from the amine group of Asp C2 and C8 originate from formate N3 and N9 are contributed by the amide group of Gln C4, C5 and N7 are derived from Gly C6 comes from HCO3- (CO2) |

The inosinate synthesis is complex, beginning with a 5-phosphoribosyl-1-pyrophosphate (PRPP). In the first step, an amino group given by glutamine is attached at carbon 1 of PRPP. The resulting molecule is 5-phosphoribosylamine, which is highly unstable, with a half-life of 30 seconds at physiologic pH. 5-Phosphoribosylamine gains an amino acid (glycine), becoming glycinamide ribonucleotide (GAR). Then, N10-formyltetrahydrofolate (THF) transfers a formyl group to glycinamide ribonucleotide to form formyl glycinamide ribonucleotide (FGAR)[4].
Using an ATP molecule, ammonia is added to the compound to become formylglycinamidine ribonucleotide. Another ATP molecule causes an intermolecular reaction that produces an imidazole ring (5-aminoimidazole ribonucleotide).
The next step of the pathway is adding bicarbonate to make carboxyaminoimidazole ribonucleotide by using ATP (it only happens in fungi and bacteria; high eukaryotes simply add CO2 to form the ribonucleotide). Then, the imidazole’s carboxylate group phosphatises and adds aspartate.
As we have just seen, a six-step process links glycine, formate, bicarbonate, glutamine, and aspartate to lead to an intermediate that contains almost all the required atoms to synthesize a purine ring. This intermediate removes fumarate, and a second formyl group from THF is added. The compound gets cycled and forms inosinate after a sort of intermolecular reactions. Inosinate is the first intermediate in this synthesis pathway to have a whole purine ring.
Enzymes taking part in IMP synthesis constitute a multienzyme complex in the cell. Evidences demonstrate that there are multifunctional enzymes, and some of them catalyze non-sequential steps in the pathway[5].
Laboratory synthesis of purine
[edit | edit source]History
[edit | edit source]The name 'purine' (purum uricum) was coined by the German chemist Emil Fischer in 1884. He synthesized it for the first time in 1899.[6] The starting material for the reaction sequence was uric acid (8), which had been isolated from kidney stones by Scheele in 1776.[7] Uric acid (8) was reacted with PCl5 to give 2,6,8-trichloropurine (10), which was converted with HI and PH4I to give 2,6-diiodopurine (11). The product was reduced to purine (1) using zinc-dust.
In addition to in vivo synthesis of purines in purine metabolism, purine can also be created artificially.
Purine (1) is obtained in good yield when formamide is heated in an open vessel at 170 oC for 28 hours.[8]
This remarkable reaction and others like it have been discussed in the context of the origin of life.[9]
Procedure:[8] Formamide (45 grams) was heated in an open vessel with a condenser for 28 hours in an oil bath at 170-190 oC. After removing excess formamide (32.1 grams) by vacuum distillation, the residue was refluxed with methanol. The methanol solvent was filtered, the solvent removed from the filtrate by vacuum distillation, and almost pure purine obtained; yield 4.93 grams (71% yield from formamide consumed). Crystallization from acetone afforded purine as colorless crystals; melting point 218 oC.
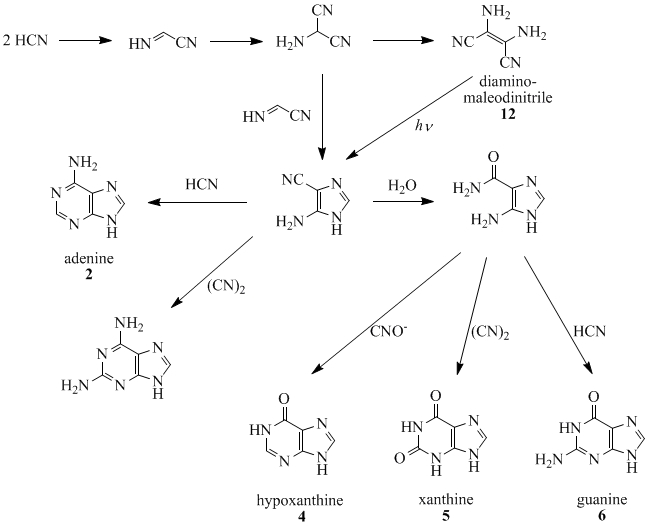
Oro, Orgel and co-workers have shown that four molecules of HCN tetramerize to form diaminomaleodinitrile (12), which can be converted into almost all natural-occurring purines.[10][11][12][13][14]
The Traube purine synthesis (1900) is a classic reaction (named after Wilhelm Traube) between an amine-substituted pyrimidine and formic acid.[15]
De novo biosynthesis of pyrimidine
[edit | edit source]Unlike purines, pyrimidines are assembled before being attached to 5-phosphoribosyl-1-pyrophosphate (PRPP).
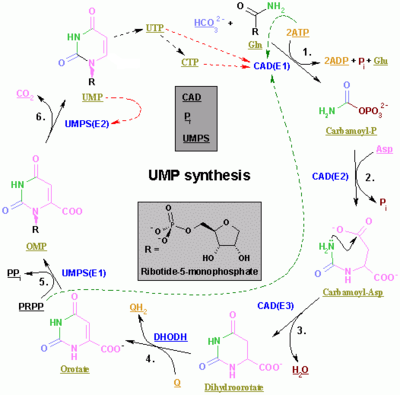
| Enzyme | Product | Description |
| carbamoyl phosphate synthetase II[16] | carbamoyl phosphate | This is the regulated step in the pyrimidine biosynthesis. |
| aspartic transcarbamolyase (aspartate carbamoyl transferase)[17] | carbamoyl aspartic acid | - |
| dihhydroorotase[18] | dihydroorotate | Dehydration |
| dihydroorotate dehydrogenase[19] (the only mitochondrial enzyme) | orotate | Dihydroorotate then enters the mitochondria where it is oxidised through removal of hydrogens. This is the only mitochondrial step in nucleotide rings biosynthesis. |
| orotate phosphoribosyltransferase[20] | OMP | PRPP is used. |
| OMP decarboxylase[21] | UMP | Decarboxylation |
| uridine-cytidine kinase 2[22] | UDP | Phosphorylation. ATP is used. |
| nucleoside diphosphate kinase | uridine 5'-triphosphate(UTP) | Phosphorylation. ATP is used. |
| CTP synthase | cytidine 5'triphosphate(CTP) | Glutamine and ATP are used. |
The first three enzymes are all coded by the same gene in Metazoa (CAD). In Fungi, a similar protein exists but lacks the dihydroorotase function: another protein catalyzes the second step.
In other organisms (Bacteria, Archaea and the other Eukaryota]]), the first three steps are done by three different enzymes.
CTP synthase (or CTP synthetase) is an enzyme involved in pyrimidine biosynthesis. It intraconverts UTP and CTP. The source of the amine/amino group in CTP is glutamine. CTP synthase is activated by GTP, a purine. This acts to balance the relative amounts of purine and pyrimidine nucleotides. CTP synthase is inhibited by reversible by CTP and irreversible for example by the glutamine analogon DON. The following human genes encode proteins that possess CTP synthase activity:
CTPS – CTP synthase 1
CTPS2 – CTP synthase 2 Pyrimidine synthesis inhibitors are used in active moderate to severe rheumatoid arthritis and psoriatic arthritis. Examples include Leflunomide and Teriflunomide.
Chemical synthesis
[edit | edit source]Pyrimidines can also be prepared within the laboratory by organic synthesis. One method is the classic Biginelli reaction. Many other methods rely on condensation of carbonyls with amines for instance the synthesis of 2-Thio-6-methyluracil from thiourea and ethyl acetoacetate [23] or the synthesis of 4-methylpyrimidine with 4,4-dimethoxy-2-butanone and formamide.[24][25]
A novel method is by reaction of certain amides with carbonitriles under electrophilic activation of the amide with 2-chloro-pyridine and trifluoromethanesulfonic anhydride [26]:
Salvage pathway
[edit | edit source]A salvage pathway is a Metabolic pathway|pathway in which nucleotides (purine and pyrimidine) are synthesized from intermediates in the degradative pathway for nucleotides.
Salvage pathways are used to recover bases and nucleosides that are formed during degradation of RNA and DNA. This is important in some organs because some tissues cannot undergo de novo synthesis.
The salvaged bases and nucleosides can then be converted back into nucleotides[27].
Substrates
[edit | edit source]The salvage pathway requires distinct substrates:
Pyrimidines
[edit | edit source]Uridine phosphorylase adds ribose-1-phospate to the free base uracil, forming uridine monophosphate. Uridine kinase then phosphorylates this nucleoside into its diphosphate and triphosphate forms. Deoxythymidine phosphorylase adds deoxyribose-1-phosphate to thymine, forming deoxythymidine monophosphate. Thymidine kinase can then phosphorylate this compound to deoxythymidine diphosphate and triphosphate.
Thymidine kinase Higher organisms have two isoenzymes, that are chemically very different, TK1 and TK2. The former was first found in fetal tissue, the second was found to be more abundant in adult tissue, and initially they were termed fetal and adult thymidine kinase. Soon it was shown that TK1 is present in the cytoplasm only in anticipation of cell division (cell cycle-dependent), whereas TK2 is located in mitochondria and is cell cycle-independent. The genes of the two types were localized in the mid-1970s. The gene for TK1 was cloned and sequenced. The corresponding protein has a molecular weight of about 25 kD. Normally, it occurs in tissue as a dimer. It can be activated by ATP. After activation, it has been converted to a tetramer. The recombinant TK1 cannot be activated and converted to a tetramer in this way, showing that the enzyme occurring in cells has been modified after synthesis. TK1 is synthesized by the cell during the S phase of cell division. After cell division is completed, TK1 is degraded intracellularly, so that it does not pass to body fluids after normal cell division. There is a feed-back regulation of the action of thymidine kinase in the cell: thymidine triphosphate (TTP), the product of the further phosphorylation of thymidine, acts as an inhibitor to thymidine kinase. This serves to maintain a balanced amount of TTP phosphate available for nucleic acid synthesis, not oversaturating the system. Genes for virus specific thymidine kinases have been identified in Herpes simplex virus, Varicella zoster virus and Epstein-Barr virus[28][29].
Deoxythymidine reacts with ATP to give deoxythymidine monophosphate and ADP.
Purines
[edit | edit source]Phosphoribosyltransferases add activated ribose-5-phosphate (called phosphoribosyl pyrophosphate or PRPP) to bases, creating nucleotide monophosphates. There are two types of phosphoribosyltransferases: adenosine phosphoribosyltransferase (APRT) and hypoxanthine-guanine phosphoribosyltransferase (HGPRT). Lesch-Nyhan syndrome is associated with a deficiency of HGPRT.
| Nucleoside | Enzyme | Nucleotide |
| hypoxanthine | hypoxanthine/guanine phosphoribosyl transferase (HGPRT) | IMP |
| guanine | hypoxanthine/guanine phosphoribosyl transferase (HGPRT) | GMP |
| adenine | adenine phosphoribosyltransferase (APRT) | AMP |
Refrences
[edit | edit source]- ↑ http://en.wikipedia.org/w/index.php?title=Purine_metabolism&oldid=423783946
- ↑ http://en.wikipedia.org/w/index.php?title=GMP_synthase&oldid=420670515
- ↑ Tesmer JJ, Klem TJ, Deras ML, Davisson VJ, Smith JL (January 1996). "The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families". Nat. Struct. Biol. 3 (1): 74–86.
- ↑ http://en.wikipedia.org/w/index.php?title=Inosinic_acid&oldid=423592099
- ↑ http://en.wikipedia.org/w/index.php?title=Inosinic_acid&oldid=423592099
- ↑ Fischer, E. Berichte der Deutschen Chemischen Gesellschaft 1899, 32, 2550.
- ↑ Scheele, V. Q. Examen Chemicum Calculi Urinari, Opuscula, 1776, 2, 73.
- ↑ a b Yamada, H.; Okamoto, T. (1972). "A One-step Synthesis of Purine Ring from Formamide". Chemical & Pharmaceutical Bulletin. 20: 623.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Saladino; Crestini, Claudia; Ciciriello, Fabiana; Costanzo, Giovanna; Mauro, Ernesto; et al. (2006). "About a Formamide-Based Origin of Informational Polymers: Syntheses of Nucleobases and Favourable Thermodynamic Niches for Early Polymers". Origins of Life and Evolution of Biospheres. 36 (5–6): 523–531. doi:10.1007/s11084-006-9053-2. PMID 17136429.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ↑ Sanchez, R. A.; Ferris, J. P.; Orgel, L. E. Journal of Molecular Biology, 1967, 30, 223.
- ↑ Ferris, J. P.; Orgel, L. E. Journal of the American Chemical Society, 1966, 88, 1074.
- ↑ Ferris, J. P.; Kuder, J. E.; Catalano, O. W. Science, 1969, 166, 765.
- ↑ Oro, J.; Kamat, J. S. Nature, 1961, 190, 442.
- ↑ Houben-Weyl, Vol . E5, p. 1547
- ↑ Organic Syntheses Based on Name Reactions, Alfred Hassner, C. Stumer ISBN 008043259X 2002
- ↑ "Entrez Gene: CAD carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase".
- ↑ "Entrez Gene: CAD carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase".
- ↑ "Entrez Gene: CAD carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase".
- ↑ "Entrez Gene: DHODH dihydroorotate dehydrogenase".
- ↑ "Entrez Gene: UMPS uridine monophosphate synthetase".
- ↑ "Entrez Gene: UMPS uridine monophosphate synthetase".
- ↑ "Entrez Gene: UCK2 uridine-cytidine kinase 2".
- ↑ Organic Syntheses, Coll. Vol. 4, p.638 (1963); Vol. 35, p.80 (1955) Link
- ↑ Organic Syntheses, Coll. Vol. 5, p.794 (1973); Vol. 43, p.77 (1963) Link
- ↑ http://en.wikipedia.org/w/index.php?title=Pyrimidine_metabolism&oldid=391209862
- ↑ Single-Step Synthesis of Pyrimidine Derivatives Mohammad Movassaghi and Matthew D. Hill J. Am. Chem. Soc.; 2006; 128(44) pp 14254 - 14255; (Communication)
- ↑ http://en.wikipedia.org/w/index.php?title=Nucleotide_salvage&oldid=414293528
- ↑ http://en.wikipedia.org/w/index.php?title=Thymidine_kinase&oldid=422438046
- ↑ Wintersberger E (February 1997). "Regulation and biological function of thymidine kinase". Biochem. Soc. Trans. 25 (1): 303–8.