Principles of Biochemistry/Nucleic acid I: DNA and its nucleotides
DNA is a long polymer made from repeating units called nucleotides. As first discovered by James D. Watson and Francis Crick, the structure of DNA of all species comprises two helical chains each coiled round the same axis, and each with a pitch of 34 Ångströms (3.4 nanometres) and a radius of 10 Ångströms (1.0 nanometres). According to another study, when measured in a particular solution, the DNA chain measured 22 to 26 Ångströms wide (2.2 to 2.6 nanometres), and one nucleotide unit measured 3.3 Å (0.33 nm) long. Although each individual repeating unit is very small, DNA polymers can be very large molecules containing millions of nucleotides. For instance, the largest human chromosome, chromosome number 1, is approximately 220 million base pairs long. In living organisms, DNA does not usually exist as a single molecule, but instead as a pair of molecules that are held tightly together. These two long strands entwine like vines, in the shape of a double helix. The nucleotide repeats contain both the segment of the backbone of the molecule, which holds the chain together, and a base, which interacts with the other DNA strand in the helix. A base linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. If multiple nucleotides are linked together, as in DNA, this polymer is called a polynucleotide. The backbone of the DNA strand is made from alternating phosphate and sugar residues. The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These asymmetric bonds mean a strand of DNA has a direction. In a double helix the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are antiparallel. The asymmetric ends of DNA strands are called the 5′ (five prime) and 3′ (three prime) ends, with the 5' end having a terminal phosphate group and the 3' end a terminal hydroxyl group. One major difference between DNA and RNA is the sugar, with the 2-deoxyribose in DNA being replaced by the alternative pentose sugar ribose in RNA.[1][2]
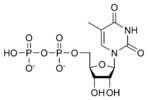
The DNA double helix is stabilized primarily by two forces: hydrogen bonds between nucleotides and base-stacking interactions among the aromatic bases. In the aqueous environment of the cell, the conjugated π bonds of nucleotide bases align perpendicular to the axis of the DNA molecule, minimizing their interaction with the solvation shell and therefore, the Gibbs free energy. The four bases found in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). These four bases are attached to the sugar/phosphate to form the complete nucleotide, as shown for adenosine monophosphate. These bases are classified into two types; adenine and guanine are fused five- and six-membered heterocyclic compounds called purines, while cytosine and thymine are six-membered rings called pyrimidines. A fifth pyrimidine base, called uracil (U), usually takes the place of thymine in RNA and differs from thymine by lacking a methyl group on its ring. Uracil is not usually found in DNA, occurring only as a breakdown product of cytosine. In addition to RNA and DNA, a large number of artificial nucleic acid analogues have also been created to study the proprieties of nucleic acids, or for use in biotechnology.[3][4]

DNA is a genetic material
[edit | edit source]
Griffith's experiment
[edit | edit source]Griffith's experiment was conducted in 1928 by Frederick Griffith, one of the first experiments suggesting that bacteria are capable of transferring genetic information through a process known as transformation. Griffith used two strains of Streptococcus pneumoniae bacteria which infect mice – a type III-S (smooth) and type II-R (rough) strain. The III-S strain covers itself with a polysaccharide capsule that protects it from the host's immune system, resulting in the death of the host, while the II-R strain doesn't have that protective capsule and is defeated by the host's immune system. A German bacteriologist, Fred Neufeld, had discovered the three pneumococcal types (Types I, II, and III) and discovered the Quellung reaction to identify them in vitro. Until Griffith's experiment, bacteriologists believed that the types were fixed and unchangeable, from one generation to another. In this experiment, bacteria from the III-S strain were killed by heat, and their remains were added to II-R strain bacteria. While neither alone harmed the mice, the combination was able to kill its host. Griffith was also able to isolate both live II-R and live III-S strains of pneumococcus from the blood of these dead mice. Griffith concluded that the type II-R had been "transformed" into the lethal III-S strain by a "transforming principle" that was somehow part of the dead III-S strain bacteria. Today, we know that the "transforming principle" Griffith observed was the DNA of the III-S strain bacteria. While the bacteria had been killed, the DNA had survived the heating process and was taken up by the II-R strain bacteria. The III-S strain DNA contains the genes that form the protective polysaccharide capsule. Equipped with this gene, the former II-R strain bacteria were now protected from the host's immune system and could kill the host. The exact nature of the transforming principle (DNA) was verified in the experiments done by Avery, McLeod and McCarty and by Hershey and Chase.[5]}}
Hershey Chase experiments
[edit | edit source]Alfred Hershey and Martha Chase conducted series of experiments in 1952 by , confirming that DNA was the genetic material, which had first been demonstrated in the 1944 Avery–MacLeod–McCarty experiment. These experiments are known as Hershey Chase experiments. The existence of DNA was known to biologists since 1869, most of them assumed that proteins carried the information for inheritance that time. Hershey and Chase conducted their experiments on the T2 phage. The phage consists of a protein shell containing its genetic material. The phage infects a bacterium by attaching to its outer membrane and injecting its genetic material and leaving its empty shell attached to the bacterium.
In their first set of experiments, Hershey and Chase labeled the DNA of phages with radioactive Phosphorus-32 (p32) (the element phosphorus is present in DNA but not present in any of the 20 amino acids which are component of proteins). They allowed the phages to infect E. coli, and through several elegant experiments were able to observe the transfer of P32 labeled phage DNA into the cytoplasm of the bacterium. In their second set of experiments, they labeled the phages with radioactive Sulfur-35 (Sulfur is present in the amino acids cysteine and methionine, but not in DNA). Following infection of E. coli they then sheared the viral protein shells off of infected cells using a high-speed blender and separated the cells and viral coats by using a centrifuge. After separation, the radioactive S35 tracer was observed in the protein shells, but not in the infected bacteria, supporting the hypothesis that the genetic material which infects the bacteria was DNA and not protein.[6][7] Hershey shared the 1969 Nobel Prize in Physiology or Medicine for his “discoveries concerning the genetic structure of viruses.”
Oswald T. Avery, Colin MacCleod, Maclyn McCarty with Francis Crick and James D Watson [8]
Bases of Nucleic acid
[edit | edit source]

Two helical strands form the DNA backbone. Another double helix may be found by tracing the spaces, or grooves, between the strands. These voids are adjacent to the base pairs and may provide a binding site. As the strands are not directly opposite each other, the grooves are unequally sized. One groove, the major groove, is 22 Å wide and the other, the minor groove, is 12 Å wide. The narrowness of the minor groove means that the edges of the bases are more accessible in the major groove. As a result, proteins like transcription factors that can bind to specific sequences in double-stranded DNA usually make contacts to the sides of the bases exposed in the major groove. This situation varies in unusual conformations of DNA within the cell, but the major and minor grooves are always named to reflect the differences in size that would be seen if the DNA is twisted back into the ordinary B form[9].
Base pairing Of DNA
[edit | edit source]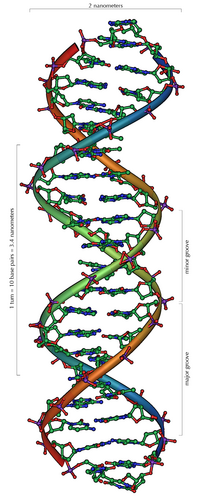
Chargaff's rules was given by Erwin Chargaff which state that DNA from any cell of all organisms should have a 1:1 ratio of pyrimidine and purine bases and, more specifically, that the amount of guanine is equal to cytosine and the amount of adenine is equal to thymine. This pattern is found in both strands of the DNA. They were discovered by Austrian chemist Erwin Chargaff.
In molecular biology, two nucleotides on opposite complementary DNA strands that are connected via hydrogen bonds are called a base pair (often abbreviated bp). In the canonical Watson-Crick DNA base pairing, Adenine (A) forms a base pair with Thymine (T) and Guanine (G) forms a base pair with Cytosine (C). In RNA, thymine is replaced by Uracil (U). Alternate hydrogen bonding patterns, such as the wobble base pair and Hoogsteen base pair, also occur—particularly in RNA—giving rise to complex and functional tertiary structures. [10]
Example
5'CTCGTTTGCGCTCTATCG3' 3'GAGCAAACGCGAGATAGC5'
Purine base
[edit | edit source]The German chemist Emil Fischer in 1884 gave the name 'purine' (purum uricum). He synthesized it for the first time in 1899 by uric acid which had been isolated from kidney stones by Scheele in 1776. Beside from DNA and RNA, purines are also components in a number of other important biomolecules, such as ATP, GTP, cyclic AMP, NADH, and coenzyme A. Purine itself, has not been found in nature, but it can be produced by organic synthesis.A purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring.
Example:
Adenine
[edit | edit source]Adenine is one of the two purine nucleobases (the other being guanine) used in forming nucleotides of the nucleic acids (DNA or RNA). In DNA, adenine binds to thymine via two hydrogen bonds to assist in stabilizing the nucleic acid structures. Adenine forms adenosine, a nucleoside, when attached to ribose, and deoxyadenosine when attached to deoxyribose. It forms adenosine triphosphate (ATP), a nucleotide, when three phosphate groups are added to adenosine.
Guanine
[edit | edit source]Guanine, along with adenine and cytosine, is present in both DNA and RNA, whereas thymine is usually seen only in DNA, and uracil only in RNA. In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with conjugated double bonds.
Guanine has two tautomeric forms, the major keto form and rare enol form. It binds to cytosine through three hydrogen bonds. In cytosine, the amino group acts as the hydrogen donor and the C-2 carbonyl and the N-3 amine as the hydrogen-bond acceptors. Guanine has a group at C-6 that acts as the hydrogen acceptor, while the group at N-1 and the amino group at C-2 act as the hydrogen donors.
Pyrimidine base
[edit | edit source]


Pyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring. It is isomeric with two other forms of diazine.Three nucleobases found in nucleic acids, cytosine (C), thymine (T), and uracil (U), are pyrimidine derivatives.
A pyrimidine has many properties in common with pyridine, as the number of nitrogen atoms in the ring increases the ring pi electrons become less energetic and electrophilic aromatic substitution gets more difficult while nucleophilic aromatic substitution gets easier. An example of the last reaction type is the displacement of the amino group in 2-aminopyrimidine by chlorine and its reverse. Reduction in resonance stabilization of pyrimidines may lead to addition and ring cleavage reactions rather than substitutions. One such manifestation is observed in the Dimroth rearrangement. Compared to pyridine, N-alkylation and N-oxidation is more difficult, and pyrimidines are also less basic: The pKa value for protonated pyrimidine is 1.23 compared to 5.30 for pyridine.[11] Pyrimidine also is found in meteorites, although scientists still do not know its origin. Pyrimidine also photolytically decomposes into Uracil under UV light.

Cytosine
[edit | edit source]Cytosine can be found as part of DNA, as part of RNA, or as a part of a nucleotide. As cytidine triphosphate (CTP), it can act as a co-factor to enzymes, and can transfer a phosphate to convert adenosine diphosphate (ADP) to adenosine triphosphate (ATP).The nucleoside of cytosine is cytidine. In DNA and RNA, cytosine is paired with guanine. However, it is inherently unstable, and can change into uracil (spontaneous deamination). This can lead to a point mutation if not repaired by the DNA repair enzymes such as uracil glycosylase, which cleaves a uracil in DNA.
Cytosine can also be methylated into 5-methylcytosine by an enzyme called DNA methyltransferase or be methylated and hydroxylated to make 5-hydroxymethylcytosine. Active enzymatic deamination of cytosine or 5-methylcytosine by the APOBEC family of cytosine deaminases could have both beneficial and detrimental implications on various cellular processes as well as on organismal evolution. The implications of deamination on 5-hydroxymethylcytosine, on the other hand, remains less understood.[12]
Thymine
[edit | edit source]Thymine (T, Thy) is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at the 5th carbon. In RNA, thymine is replaced with uracil in most cases. In DNA, thymine(T) binds to adenine (A) via two hydrogen bonds, thus stabilizing the nucleic acid structures.
Uracil
[edit | edit source]Uracil found in RNA, it base-pairs with adenine and replaces thymine during DNA transcription. Methylation of uracil produces thymine. It turns into thymine to protect the DNA and to improve the efficiency of DNA replication. Uracil can base-pair with any of the bases, depending on how the molecule arranges itself on the helix, but readily pairs with adenine because the methyl group is repelled into a fixed position. Uracil pairs with adenine through hydrogen bonding. Uracil is the hydrogen bond acceptor and can form two hydrogen bonds. Uracil can also bind with a ribose sugar to form the ribonucleoside uridine. When a phosphate attaches to uridine, uridine 5'-monophosphate is produced.
Nucleosides and Nucleotides
[edit | edit source]Nucleotides
[edit | edit source]| Nitrogenous base | Nucleoside | Deoxynucleoside |
|---|---|---|
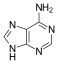 w:Adenine |
 w:Adenosine A |
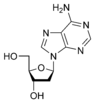 w:Deoxyadenosine dA |
 w:Guanine |
 w:Guanosine G |
 w:Deoxyguanosine dG |
 w:Thymine |
 w:5-Methyluridine m5U |
 w:Thymidine dT |
 w:Uracil |
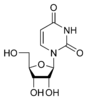 w:Uridine U |
 w:Deoxyuridine dU |
 w:Cytosine |
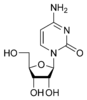 w:Cytidine C |
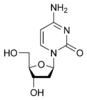 w:Deoxycytidine dC |
Nucleosides are glycosylamines consisting of a nucleobase (often referred to as simply base) bound to a ribose or deoxyribose sugar via a beta-glycosidic linkage. Examples of nucleosides include cytidine, uridine, adenosine, guanosine, thymidine and inosine. Nucleosides can be phosphorylated by specific kinases in the cell on the sugar's primary alcohol group (-CH2-OH), producing nucleotides, which are the molecular building-blocks of DNA and RNA[13].
Nucleosides can be produced by de novo synthesis pathways, in particular in the liver, but they are more abundantly supplied via ingestion and digestion of nucleic acids in the diet, whereby nucleotidases break down nucleotides (such as the thymine nucleotide) into nucleosides (such as thymidine) and phosphate.
1. Adenosine is a nucleoside composed of a molecule of adenine attached to a ribose sugar molecule (ribofuranose) moiety via a β-N9-glycosidic bond.
2. Cytidine is a nucleoside molecule that is formed when cytosine is attached to a ribose ring (also known as a ribofuranose) via a β-N1-glycosidic bond. Cytidine is a component of RNA.
3. Guanosine is a purine nucleoside comprising guanine attached to a ribose (ribofuranose) ring via a β-N9-glycosidic bond. Guanosine can be phosphorylated to become guanosine monophosphate (GMP), cyclic guanosine monophosphate (cGMP), guanosine diphosphate (GDP), and guanosine triphosphate (GTP).
4. Thymidine (more precisely called deoxythymidine; can also be labelled deoxyribosylthymine, and thymine deoxyriboside) is a chemical compound, more precisely a pyrimidine deoxynucleoside. Deoxythymidine is the DNA nucleoside T, which pairs with deoxyadenosine (A) in double-stranded DNA.
If cytosine is attached to a deoxyribose ring, it is known as a deoxycytidine.[14]
Nucleotide
[edit | edit source]
A nucleotide is composed of a nucleobase (nitrogenous base), a five-carbon sugar (either ribose or 2'-deoxyribose), and one to three phosphate groups. Together, the nucleobase and sugar comprise a nucleoside. The phosphate groups form bonds with either the 2, 3, or 5-carbon of the sugar, with the 5-carbon site most common. Cyclic nucleotides form when the phosphate group is bound to two of the sugar's hydroxyl groups. Ribonucleotides are nucleotides where the sugar is ribose, and deoxyribonucleotides contain the sugar deoxyribose. Nucleotides can contain either a purine or a pyrimidine base. Nucleic acids are polymeric macromolecules made from nucleotide monomers. In DNA, the purine bases are adenine and guanine, while the pyrimidines are thymine and cytosine. RNA uses uracil in place of thymine. Adenine always pairs with thymine by 2 hydrogen bonds, while guanine pairs with cytosine through 3 hydrogen bonds, each due to their unique structures.[15]
A deoxyribonucleotide is the monomer, or single unit, of DNA, or deoxyribonucleic acid. Each deoxyribonucleotide comprises three parts: a nitrogenous base, a deoxyribose sugar, and one or more phosphate groups. The nitrogenous base is always bonded to the 1' carbon of the deoxyribose, which is distinguished from ribose by the presence of a proton on the 2' carbon rather than an -OH group. The phosphate groups bind to the 5' carbon of the sugar. When deoxyribonucleotides polymerize to form DNA, the phosphate group from one nucleotide will bond to the 3' carbon on another nucleotide, forming a phosphodiester bond via dehydration synthesis. New nucleotides are always added to the 3' carbon of the last nucleotide, so synthesis always proceeds from 5' to 3'[16].
Phosphodiester bond
A phosphodiester bond is a group of strong covalent bonds between a phosphate group and two 5-carbon ring carbohydrates (pentoses) over two ester bonds. Phosphodiester bonds are central to most life on Earth, as they make up the backbone of the strands of DNA. In DNA and RNA, the phosphodiester bond is the linkage between the 3' carbon atom of one sugar molecule and the 5' carbon of another, deoxyribose in DNA and ribose in RNA. The phosphate groups in the phosphodiester bond are negatively-charged. Because the phosphate groups have a pKa near 0, they are negatively-charged at pH 7. This repulsion forces the phosphates to take opposite sides of the DNA strands and is neutralized by proteins (histones), metal ions such as magnesium, and polyamines. In order for the phosphodiester bond to be formed and the nucleotides to be joined, the tri-phosphate or di-phosphate forms of the nucleotide building blocks are broken apart to give off energy required to drive the enzyme-catalyzed reaction. When a single phosphate or two phosphates known as pyrophosphates break away and catalyze the reaction, the phosphodiester bond is formed. Hydrolysis of phosphodiester bonds can be catalyzed by the action of phosphodiesterases which play an important role in repairing DNA sequences. In biological systems, the phosphodiester bond between two ribonucleotides can be broken by alkaline hydrolysis because of the free 2' hydroxyl group[17].

DNA structure determination using molecular modeling and DNA X-ray patterns
[edit | edit source]After DNA has been separated and purified by standard biochemical techniques one has a sample in a jar much like in the figure at the top of this article. Below are the main steps involved in generating structural information from X-ray diffraction studies of oriented DNA fibers that are drawn from the hydrated DNA sample with the help of molecular models of DNA that are combined with crystallographic and mathematical analysis of the X-ray patterns.
Paracrystalline lattice models of B-DNA structures
[edit | edit source]
A paracrystalline lattice, or paracrystal, is a molecular or atomic lattice with significant amounts (e.g., larger than a few percent) of partial disordering of molecular arrangements. Limiting cases of the paracrystal model are nanostructures, such as glasses, liquids, etc., that may possess only local ordering and no global order. A simple example of a paracrystalline lattice is shown in the following figure for a silica glass:
Liquid crystals also have paracrystalline rather than crystalline structures.
Highly hydrated B-DNA occurs naturally in living cells in such a paracrystalline state, which is a dynamic one in spite of the relatively rigid DNA double-helix stabilized by parallel hydrogen bonds between the nucleotide base-pairs in the two complementary, helical DNA chains (see figures). For simplicity most DNA molecular models omit both water and ions dynamically bound to B-DNA, and are thus less useful for understanding the dynamic behaviors of B-DNA in vivo. The physical and mathematical analysis of X-ray[19][20] and spectroscopic data for paracrystalline B-DNA is therefore much more complicated than that of crystalline, A-DNA X-ray diffraction patterns. The paracrystal model is also important for DNA technological applications such as DNA nanotechnology. Novel techniques that combine X-ray diffraction of DNA with X-ray microscopy in hydrated living cells are now also being developed.[21]
Forms of DNA
[edit | edit source]A-DNA: A-DNA is one of the many possible double helical structures of DNA. A-DNA is thought to be one of three biologically active double helical structures along with B- and Z-DNA. It is a right-handed double helix fairly similar to the more common and well-known B-DNA form, but with a shorter more compact helical structure. It appears likely that it occurs only in dehydrated samples of DNA, such as those used in crystallographic experiments, and possibly is also assumed by DNA-RNA hybrid helices and by regions of double-stranded RNA[22].
B-DNAThe most common form of DNA is B-DNA. The DNA double helix is a spiral polymer of nucleic acids, held together by nucleotides which base pair together. In B-DNA, the most common double helical structure, the double helix is right-handed with about 10–10.5 nucleotides per turn. The double helix structure of DNA contains a major groove and minor groove, the major groove being wider than the minor groove. Given the difference in widths of the major groove and minor groove, many proteins which bind to DNA do so through the wider major groove. The geometry of a base, or base pair step can be characterized by 6 coordinates: Shift, slide, rise, tilt, roll, and twist. These values precisely define the location and orientation in space of every base or base pair in a nucleic acid molecule relative to its predecessor along the axis of the helix. Together, they characterize the helical structure of the molecule. In regions of DNA or RNA where the "normal" structure is disrupted, the change in these values can be used to describe such disruption. For each base pair, considered relative to its predecessor, there are the following base pair geometries to consider:
Shear: bases appear as though they have moved past one another on the xy-plane.
Stretch: two bases are stretched apart in the horizontal direction.
Stagger: bases have both moved past one another and stretched apart on the xy-plane.
Buckle: the ends of the bases that do not touch are inclined upwards towards the z-axis.
Propeller twist: rotation of one base with respect to the other in the same base pair.
Opening: bases touch at one side, but lean apart at the other side.
Shift: displacement along an axis in the base-pair plane perpendicular to the first, directed from the minor to the major groove.
Slide: displacement along an axis in the plane of the base pair directed from one strand to the other.
Rise: displacement along the helix axis.
Tilt: rotation around this axis.
vRoll: rotation around this axis.
Twist: rotation around the helix axis.
vx-displacement
y-displacement
inclination
tip pitch: the number of base pairs per complete turn of the helix.

Rise and twist determine the handedness and pitch of the helix. The other coordinates, by contrast, can be zero. Slide and shift are typically small in B-DNA, but are substantial in A- and Z-DNA. Roll and tilt make successive base pairs less parallel, and are typically small. A diagram of these coordinates can be found in 3DNA website. Note that "tilt" has often been used differently in the scientific literature, referring to the deviation of the first, inter-strand base-pair axis from perpendicularity to the helix axis. This corresponds to slide between a succession of base pairs, and in helix-based coordinates is properly termed "inclination".
TA-DNA: TA-DNA is a form of DNA that is most closely related to A-DNA in terms of structure. The TA-DNA helix is right handed like A-DNA, and it The most noticeable difference between the two forms is that TA-DNA has a greater inclination of its base pairs at an angle of approximately 50 degrees with respect to the helix axis. Because of this, TA-DNA is sometimes described as tilted A-DNA.[23]
Z-DNA: Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the double helix winds to the left in a zig-zag pattern (instead of to the right, like the more common B-DNA form). Z-DNA is thought to be one of three biologically active double helical structures along with A- and B-DNA. Z-DNA is quite different from the right-handed forms. In fact, Z-DNA is often compared against B-DNA in order to illustrate the major differences. The Z-DNA helix is left-handed and has a structure that repeats every 2 base pairs. The major and minor grooves, unlike A- and B-DNA, show little difference in width. Formation of this structure is generally unfavourable, although certain conditions can promote it; such as alternating purine-pyrimidine sequence (especially poly(dGC)2), negative DNA supercoiling or high salt and some cations (all at physiological temperature, 37°C, and pH 7.3-7.4). Z-DNA can form a junction with B-DNA (called a "B-to-Z junction box") in a structure which involves the extrusion of a base pair. The Z-DNA conformation has been difficult to study because it does not exist as a stable feature of the double helix. Instead, it is a transient structure that is occasionally induced by biological activity and then quickly disappears.[24][25]
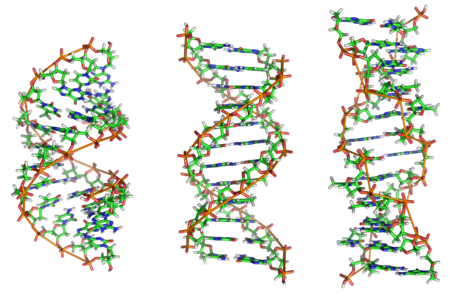
| A-DNA | B-DNA | Z-DNA | |
|---|---|---|---|
| Helix sense | Right-handed | Right-handed | Left-handed |
| Diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
| Repeating unit | 1 bp | 1 bp | 2 bp |
| Rotation/bp | 32.7° | 35.9° | 60°/2 |
| bp/turn | 11 | 10.5 | 12 |
| Inclination of bp to axis | +19° | −1.2° | −9° |
| Rise/bp along axis | 2.3 Å (0.23 nm) | 3.32 Å (0.332 nm) | 3.8 Å (0.38 nm) |
| Pitch/turn of helix | 28.2 Å (2.82 nm) | 33.2 Å (3.32 nm) | 45.6 Å (4.56 nm) |
| Mean propeller twist | +18° | +16° | 0° |
| Glycosyl angle | anti | anti | C: anti, G: syn |
| Sugar pucker | C3'-endo | C2'-endo | C: C2'-endo, G: C2'-exo |
bp-Base pair, nm-nano meter
Minor and Major Grooves of DNA: The minor and major grooves of DNA are characteristic of the type of DNA in which they are a part of. They are very important because they not only make up the characteristic structure of A-DNA, B-DNA, and Z-DNA, but their differences determine the interaction of these DNA with various proteins. The wideness, narrowness, deepness, shallowness, and the electrostatic potential found within these grooves all play an important role in determining how the DNA will react with certain proteins, with which proteins it will react, and whether or not certain parts will be likely to react at all.[26]
| Minor Groove | Major Groove | |
|---|---|---|
| A-DNA | Wide and Shallow with relatively lower electrostatic potential | Narrow and Deep with relatively high electrostatic potential |
| B-DNA | Narrow and Deep with relatively high electrostatic potential | Wide and Shallow with relatively lower electrostatic potential |
| Z-DNA | Narrow and Deep the a zigzag curving of the backbone | Indistinct and the base edges create a convex surface |

Differences in the Relative Charges of A-DNA, B-DNA, and Z-DNA: For all forms of DNA the global structure has an overall negative charge and the overall electrostatic potential is negative. This is largely due to the phosphate groups of DNA which bear an overall negative charge due to the negatively charged oxygen atoms they have. However, DNA has both positive and negative charges covering its outer structure. The charge dispersion over the global structure of DNA varies between A-DNA, B-DNA, and Z-DNA. A-DNA has mostly negative electrostatic potential throughout but concentrated mostly in the site of the major groove. It also has neutral electrostatic potential spread widely throughout the rest of its structure besides the major groove. Finally, A-DNA has sparse positive charges scattered along its surface. B-DNA has mostly negative electrostatic potential spread throughout its global exterior and concentrated mostly in major and minor grooves. It has some positively charged surface area scattered sparsely over its surface and mostly peppered within the wide major groove. Z-DNA has a negatively charged minor groove and the indistinct area where a major groove would normally be has slight positive charged places. Overall, the Z-DNA has a negative charge in all of its global structure. The structures of A-DNA, B-DNA, and Z-DNA have an overall negative charge. [27]
| A-DNA | B-DNA | Z-DNA | |
|---|---|---|---|
| Negative Potential | Massively present throughout the global structure but concentrated most highly in the major groove | Massively present throughout the global structure and evenly concentrated in the major and minor grooves | Massively present throughout the global structure and present most highly in the minor groove |
| Positive Potential | Very sparsely present scattered in the minor groove and hardly at all in the major groove | Very sparsely present scattered throughout the global structure, mostly in the major groove | Scattered sparsely in greater amounts than in A-DNA or B-DNA, but outside of the minor groove |
| Neutral Potential | Moderately present throughout the structure and mostly in the minor groove, but not in the major groove | Moderately scattered throughout the structure, but in a smaller amount than in A-DNA | Moderately scattered throughout the entire global structure apart from the minor groove |

Coiling of DNA
[edit | edit source]

DNA supercoiling is important for DNA packaging within all cells. Because the length of DNA can be thousands of times that of a cell, packaging this genetic material into the cell or nucleus (in eukaryotes) is a difficult feat. Supercoiling of DNA reduces the space and allows for a lot more DNA to be packaged. In prokaryotes, plectonemic supercoils are predominant, because of the circular chromosome and relatively small amount of genetic material. In eukaryotes, DNA supercoiling exists on many levels of both plectonemic and solenoidal supercoils, with the solenoidal supercoiling proving most effective in compacting the DNA. Solenoidal supercoiling is achieved with histones to form a 10nm fiber. This fiber is further coiled into a 30nm fiber, and further coiled upon itself numerous times more. DNA packaging is greatly increased during nuclear division events such as mitosis or meiosis, where DNA must be compacted and segregated to daughter cells. Condensins and cohesins are Structural Maintenance of Chromosome proteins that aid in the condensation of sister chromatids and the linkage of the centromere in sister chromatids. These SMC proteins induce positive supercoils. Supercoiling is also required for DNA/RNA synthesis. Because DNA must be unwound for DNA/RNA polymerase action, supercoils will result. The region ahead of the polymerase complex will be unwound; this stress is compensated with positive supercoils ahead of the complex. Behind the complex, DNA is rewound and there will be compensatory negative supercoils. It is important to note that topoisomerases such as DNA gyrase (Type II Topoisomerase) play a role in relieving some of the stress during DNA/RNA synthesis[28].
NA supercoiling can be described numerically by changes in the 'linking number' Lk. The linking number is the most descriptive property of supercoiled DNA. Lko, the number of turns in the relaxed (B type) DNA plasmid/molecule, is determined by dividing the total base pairs of the molecule by the relaxed bp/turn which, depending on reference is 10.4-10.5.
Lk is merely the number of crosses a single strand makes across the other in a planar projection. The topology of the DNA is described by the equation below in which the linking number is equivalent to the sum of TW, which is the number of twists or turns of the double helix, and Wr which is the number of coils or 'writhes'. If there is a closed DNA molecule, the sum of TW and Wr, or the linking number, does not change. However, there may be complementary changes in TW and Wr without changing their sum.
The change in the linking number, ΔLk, is the actual number of turns in the plasmid/molecule, Lk, minus the number of turns in the relaxed plasmid/molecule Lko.
If the DNA is negatively supercoiled ΔLk < 0. The negative supercoiling implies that the DNA is underwound.
A standard expression independent of the molecule size is the "specific linking difference" or "superhelical density" denoted σ. σ represents the number of turns added or removed relative to the total number of turns in the relaxed molecule/plasmid, indicating the level of supercoiling.
The Gibbs free energy associated with the coiling is given by the equation below[30]
The linking number is a numerical invariant that describes the linking of two closed curves in three-dimensional space. Intuitively, the linking number represents the number of times that each curve winds around the other. The linking number is always an integer, but may be positive or negative depending on the orientation of the two curves[31]. Since the linking number L of supercoiled DNA is the number of times the two strands are intertwined (and both strands remain covalently intact), L cannot change. The reference state (or parameter) L0 of a circular DNA duplex is its relaxed state. In this state, its writhe W = 0. Since L = T + W, in a relaxed state T = L. Thus, if we have a 400 bp relaxed circular DNA duplex, L ~ 40 (assuming ~10 bp per turn in B-DNA). Then T ~ 40.
- Positively supercoiling:
- T = 0, W = 0, then L = 0
- T = +3, W = 0, then L = +3
- T = +2, W = +1, then L = +3
- Negatively supercoiling:
- T = 0, W = 0, then L = 0
- T = -3, W = 0, then L = -3
- T = -2, W = -1, then L = -3
Negative supercoils favor local unwinding of the DNA, allowing processes such as transcription, DNA replication, and recombination. Negative supercoiling is also thought to favour the transition between B-DNA and Z-DNA, and moderate the interactions of DNA binding proteins involved in gene regulation.[32][33]
DNA sequencing
[edit | edit source]RNA sequencing was one of the earliest forms of nucleotide sequencing. The major landmark of RNA sequencing is the sequence of the first complete gene and the complete genome of Bacteriophage MS2, identified and published by Walter Fiers and his coworkers at the University of Ghent (Ghent, Belgium), between 1972 and 1976. Prior to the development of rapid DNA sequencing methods in the early 1970s by Frederick Sanger at the University of Cambridge, in England and Walter Gilbert and Allan Maxam at Harvard, a number of laborious methods were used. For instance, in 1973, Gilbert and Maxam reported the sequence of 24 basepairs using a method known as wandering-spot analysis. The chain-termination method developed by Sanger and coworkers in 1975 soon became the method of choice, owing to its relative ease and reliability.[34]
Maxam and Gilbert method
[edit | edit source]In 1976–1977, Allan Maxam and Walter Gilbert developed a DNA sequencing method based on chemical modification of DNA and subsequent cleavage at specific bases. Although Maxam and Gilbert published their chemical sequencing method two years after the ground-breaking paper of Sanger and Coulson on plus-minus sequencing,Maxam–Gilbert sequencing rapidly became more popular, since purified DNA could be used directly, while the initial Sanger method required that each read start be cloned for production of single-stranded DNA. However, with the improvement of the chain-termination method (see below), Maxam-Gilbert sequencing has fallen out of favour due to its technical complexity prohibiting its use in standard molecular biology kits, extensive use of hazardous chemicals, and difficulties with scale-up. The method requires radioactive labeling at one 5' end of the DNA (typically by a kinase reaction using gamma-32P ATP) and purification of the DNA fragment to be sequenced. Chemical treatment generates breaks at a small proportion of one or two of the four nucleotide bases in each of four reactions (G, A+G, C, C+T). For example, the purines (A+G) are depurinated using formic acid, the guanines (and to some extent the adenines) are methylated by dimethyl sulfate, and the pyrimidines (C+T) are methylated using hydrazine. The addition of salt (sodium chloride) to the hydrazine reaction inhibits the methylation of thymine for the C-only reaction. The modified DNAs are then cleaved by hot piperidine at the position of the modified base. The concentration of the modifying chemicals is controlled to introduce on average one modification per DNA molecule. Thus a series of labeled fragments is generated, from the radiolabeled end to the first "cut" site in each molecule. The fragments in the four reactions are electrophoresed side by side in denaturing acrylamide gels for size separation. To visualize the fragments, the gel is exposed to X-ray film for autoradiography, yielding a series of dark bands each corresponding to a radiolabeled DNA fragment, from which the sequence may be inferred. Also sometimes known as "chemical sequencing", this method led to the Methylation Interference Assay used to map DNA-binding sites for DNA-binding proteins.[35]
Dideoxynucleotide Chain-termination methods
[edit | edit source]
Because the chain-terminator method (or Sanger method after its developer Frederick Sanger) is more efficient and uses fewer toxic chemicals and lower amounts of radioactivity than the method of Maxam and Gilbert, it rapidly became the method of choice. The key principle of the Sanger method was the use of dideoxynucleotide triphosphates (ddNTPs) as DNA chain terminators.
The classical chain-termination method requires a single-stranded DNA template, a DNA primer, a DNA polymerase, normal deoxynucleotidephosphates (dNTPs), and modified nucleotides (dideoxyNTPs) that terminate DNA strand elongation. These ddNTPs will also be radioactively or fluorescently labelled for detection in automated sequencing machines. The DNA sample is divided into four separate sequencing reactions, containing all four of the standard deoxynucleotides (dATP, dGTP, dCTP and dTTP) and the DNA polymerase. To each reaction is added only one of the four dideoxynucleotides (ddATP, ddGTP, ddCTP, or ddTTP) which are the chain-terminating nucleotides, lacking a 3'-hydroxyl (OH) group required for the formation of a phosphodiester bond between two nucleotides, thus terminating DNA strand extension and resulting in DNA fragments of varying length[36].
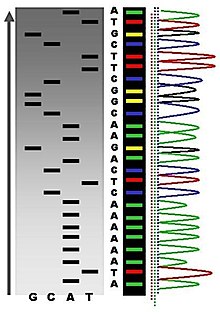
The newly synthesized and labelled DNA fragments are heat denatured, and separated by size (with a resolution of just one nucleotide) by gel electrophoresis on a denaturing polyacrylamide-urea gel with each of the four reactions run in one of four individual lanes (lanes A, T, G, C); the DNA bands are then visualized by autoradiography or UV light, and the DNA sequence can be directly read off the X-ray film or gel image. In the image on the right, X-ray film was exposed to the gel, and the dark bands correspond to DNA fragments of different lengths. A dark band in a lane indicates a DNA fragment that is the result of chain termination after incorporation of a dideoxynucleotide (ddATP, ddGTP, ddCTP, or ddTTP). The relative positions of the different bands among the four lanes are then used to read (from bottom to top) the DNA sequence[37].

Technical variations of chain-termination sequencing include tagging with nucleotides containing radioactive phosphorus for radiolabelling, or using a primer labeled at the 5’ end with a fluorescent dye. Dye-primer sequencing facilitates reading in an optical system for faster and more economical analysis and automation. The later development by Leroy Hood and coworkers [38][39] of fluorescently labeled ddNTPs and primers set the stage for automated, high-throughput DNA sequencing.
Chain-termination methods have greatly simplified DNA sequencing. For example, chain-termination-based kits are commercially available that contain the reagents needed for sequencing, pre-aliquoted and ready to use. Limitations include non-specific binding of the primer to the DNA, affecting accurate read-out of the DNA sequence, and DNA secondary structures affecting the fidelity of the sequence[40].
Dye-terminator sequencing
[edit | edit source]
Dye-terminator sequencing utilizes labelling of the chain terminator ddNTPs, which permits sequencing in a single reaction, rather than four reactions as in the labelled-primer method. In dye-terminator sequencing, each of the four dideoxynucleotide chain terminators is labelled with fluorescent dyes, each of which emit light at different wavelengths.
Owing to its greater expediency and speed, dye-terminator sequencing is now the mainstay in automated sequencing. Its limitations include dye effects due to differences in the incorporation of the dye-labelled chain terminators into the DNA fragment, resulting in unequal peak heights and shapes in the electronic DNA sequence trace chromatogram after capillary electrophoresis (see figure to the left).
This problem has been addressed with the use of modified DNA polymerase enzyme systems and dyes that minimize incorporation variability, as well as methods for eliminating "dye blobs". The dye-terminator sequencing method, along with automated high-throughput DNA sequence analyzers, is now being used for the vast majority of sequencing projects[41].
Challenges
[edit | edit source]Common challenges of DNA sequencing include poor quality in the first 15–40 bases of the sequence and deteriorating quality of sequencing traces after 700–900 bases. Base calling software typically gives an estimate of quality to aid in quality trimming.[42][43]
In cases where DNA fragments are cloned before sequencing, the resulting sequence may contain parts of the cloning vector. In contrast, PCR-based cloning and emerging sequencing technologies based on pyrosequencing often avoid using cloning vectors. Recently, one-step Sanger sequencing (combined amplification and sequencing) methods such as Ampliseq and SeqSharp have been developed that allow rapid sequencing of target genes without cloning or prior amplification.[44][45][46]
Current methods can directly sequence only relatively short (300–1000 nucleotides long) DNA fragments in a single reaction. The main obstacle to sequencing DNA fragments above this size limit is insufficient power of separation for resolving large DNA fragments that differ in length by only one nucleotide. In all cases the use of a primer with a free 5' end is essential[47].
Automation and sample preparation
[edit | edit source]
Automated DNA-sequencing instruments (DNA sequencers) can sequence up to 384 DNA samples in a single batch (run) in up to 24 runs a day. DNA sequencers carry out capillary electrophoresis for size separation, detection and recording of dye fluorescence, and data output as fluorescent peak trace chromatograms. Sequencing reactions by thermocycling, cleanup and re-suspension in a buffer solution before loading onto the sequencer are performed separately. A number of commercial and non-commercial software packages can trim low-quality DNA traces automatically. These programs score the quality of each peak and remove low-quality base peaks (generally located at the ends of the sequence). The accuracy of such algorithms is below visual examination by a human operator, but sufficient for automated processing of large sequence data sets[48].
Facts to be remembered
[edit | edit source]- 1869 DNA was first isolated by the Swiss physician Friedrich Miescher who discovered a microscopic substance in the pus of discarded surgical bandages.
- 1937 William Astbury produced the first X-ray diffraction patterns that showed that DNA had a regular structure.
- 1928 Frederick Griffith discovered that traits of the "smooth" form of the Pneumococcus could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form.
- 1952 Alfred Hershey and Martha Chase in the Hershey–Chase experiment showed that DNA is the genetic material of the T2 phage.
- 1953 James D. Watson and Francis Crick suggested double-helix model of DNA structure.
- 1972 Development of recombinant DNA technology, which permits isolation of defined fragments of DNA; prior to this, the only accessible samples for sequencing were from bacteriophage or virus DNA.
- 1977 The first complete DNA genome to be sequenced is that of bacteriophage φX174.
- 1977 Allan Maxam and Walter Gilbert publish "DNA sequencing by chemical degradation".Frederick Sanger, independently, publishes "DNA sequencing with chain-terminating inhibitors".
- 1984 Medical Research Council scientists decipher the complete DNA sequence of the Epstein-Barr virus, 170 kb.
- 1986 Leroy E. Hood's laboratory at the California Institute of Technology and Smith announce the first semi-automated DNA sequencing machine.
- 1987 Applied Biosystems markets first automated sequencing machine, the model ABI 370.
- 1990 The U.S. National Institutes of Health (NIH) begins large-scale sequencing trials on Mycoplasma capricolum, Escherichia coli, Caenorhabditis elegans, and Saccharomyces cerevisiae (at US$0.75/base).
- 1991 Sequencing of human expressed sequence tags begins in Craig Venter's lab, an attempt to capture the coding fraction of the human genome.
- 1995 Craig Venter, Hamilton Smith, and colleagues at The Institute for Genomic Research (TIGR) publish the first complete genome of a free-living organism, the bacterium Haemophilus influenzae. The circular chromosome contains 1,830,137 bases and its publication in the journal Science marks the first use of whole-genome shotgun sequencing, eliminating the need for initial mapping efforts.
- 1996 Pål Nyrén and his student Mostafa Ronaghi at the Royal Institute of Technology in Stockholm publish their method of pyrosequencing
- 1998 Phil Green and Brent Ewing of the University of Washington publish “phred” for sequencer data analysis.
- 2000 Lynx Therapeutics publishes and markets "MPSS" - a parallelized, adapter/ligation-mediated, bead-based sequencing technology, launching "next-generation" sequencing.
- 2001 A draft sequence of the human genome is published.
- 2004 454 Life Sciences markets a parallelized version of pyrosequencing. The first version of their machine reduced sequencing costs 6-fold compared to automated Sanger sequencing, and was the second of a new generation of sequencing technologies, after MPSS.
Refrences
[edit | edit source]- ↑ http://en.wikipedia.org/w/index.php?title=DNA&oldid=424928563
- ↑ Berry, Andrew; Watson, James D. (2003). DNA: the secret of life. New York: Alfred A. Knopf. ISBN 0-375-41546-7.
- ↑ http://en.wikipedia.org/w/index.php?title=DNA&oldid=424928563
- ↑ Berry, Andrew; Watson, James D. (2003). DNA: the secret of life. New York: Alfred A. Knopf. ISBN 0-375-41546-7.
- ↑ Griffith experiment
- ↑ Hershey–Chase experiment
- ↑ Hershey, A.D. and Chase, M. (1952) Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 36:39–56.
- ↑ A very–MacLeod–McCarty experiment
- ↑ http://en.wikipedia.org/w/index.php?title=DNA&oldid=427633053
- ↑ Base pair
- ↑ Pyrimidine
- ↑ Cytosine
- ↑ http://en.wikipedia.org/w/index.php?title=Nucleoside&oldid=419856059
- ↑ http://en.wikipedia.org/w/index.php?title=Nucleoside&oldid=419856059
- ↑ http://en.wikipedia.org/w/index.php?title=Nucleotide&oldid=422383586
- ↑ http://en.wikipedia.org/w/index.php?title=Nucleotide&oldid=422383586
- ↑ http://en.wikipedia.org/w/index.php?title=Phosphodiester_bond&oldid=407638927
- ↑ Herbert R. Wilson, FRS. Diffraction of X-rays by proteins, Nucleic Acids and Viruses., London: Edward Arnold (Publishers) Ltd. 1966.
- ↑ Hosemann R., Bagchi R.N., Direct analysis of diffraction by matter, North-Holland Publs., Amsterdam – New York, 1962.
- ↑ Baianu, I.C. (1978). "X-ray scattering by partially disordered membrane systems". Acta Cryst.,. A34 (5): 751–3. doi:10.1107/S0567739478001540.
{{cite journal}}: CS1 maint: extra punctuation (link) - ↑ Yamamoto Y, Shinohara K (2002). "Application of X-ray microscopy in analysis of living hydrated cells". Anat. Rec. 269 (5): 217–23. doi:10.1002/ar.10166. PMID 12379938.
{{cite journal}}: Unknown parameter|month=ignored (help) - ↑ http://en.wikipedia.org/w/index.php?title=A-DNA&oldid=379025458
- ↑ Rohs, Remo, Xiangshu Jin, Sean M. West, Rohit Joshi, Barry Honig, and Richard S. Mann. "Origins of Specificity in Protein-DNA Recognition." Annual Reviews. N.p., 24 03 2010. Web. 29 Oct 2011. <http://www.annualreviews.org/doi/full/10.1146/annurev-biochem-060408-091030?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub=pubmed>.
- ↑ http://en.wikipedia.org/w/index.php?title=Z-DNA&oldid=418927717
- ↑ Zhang H, Yu H, Ren J, Qu X (2006). "Reversible B/Z-DNA transition under the low salt condition and non-B-form polydApolydT selectivity by a cubane-like europium-L-aspartic acid complex". Biophysical Journal 90 (9): 3203–3207.
- ↑ Rohs, Remo, Xiangshu Jin, Sean M. West, Rohit Joshi, Barry Honig, and Richard S. Mann. "Origins of Specificity in Protein-DNA Recognition." Annual Reviews. N.p., 24 03 2010. Web. 29 Oct 2011. <http://www.annualreviews.org/doi/full/10.1146/annurev-biochem-060408-091030?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub=pubmed>.
- ↑ Rohs, Remo, Xiangshu Jin, Sean M. West, Rohit Joshi, Barry Honig, and Richard S. Mann. "Origins of Specificity in Protein-DNA Recognition." Annual Reviews. N.p., 24 03 2010. Web. 29 Oct 2011. <http://www.annualreviews.org/doi/full/10.1146/annurev-biochem-060408-091030?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub=pubmed>.
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_supercoil&oldid=417575151
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_supercoil&oldid=417575151
- ↑ Vologodskii AV, Lukashin AV, Anshelevich VV; et al. (1979). "Fluctuations in superhelical DNA". Nucleic Acids Res. 6: 967–682. doi:10.1093/nar/6.3.967.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ↑ http://en.wikipedia.org/w/index.php?title=DNA_supercoil&oldid=417575151
- ↑ H. S. Chawla (2002). Introduction to Plant Biotechnology. Science Publishers. ISBN 1578082285.
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_supercoil&oldid=417575151
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ Smith LM, Sanders JZ, Kaiser RJ; et al. (1986). "Fluorescence detection in automated DNA sequence analysis". Nature. 321 (6071): 674–9. doi:10.1038/321674a0. PMID 3713851.
We have developed a method for the partial automation of DNA sequence analysis. Fluorescence detection of the DNA fragments is accomplished by means of a fluorophore covalently attached to the oligonucleotide primer used in enzymatic DNA sequence analysis. A different coloured fluorophore is used for each of the reactions specific for the bases A, C, G and T. The reaction mixtures are combined and co-electrophoresed down a single polyacrylamide gel tube, the separated fluorescent bands of DNA are detected near the bottom of the tube, and the sequence information is acquired directly by computer.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ↑ Smith LM, Fung S, Hunkapiller MW, Hunkapiller TJ, Hood LE (1985). "The synthesis of oligonucleotides containing an aliphatic amino group at the 5' terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis". Nucleic Acids Res. 13 (7): 2399–412. doi:10.1093/nar/13.7.2399. PMC 341163. PMID 4000959.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ "Phred - Quality Base Calling". Retrieved 2011-02-24.
- ↑ "Base-calling for next-generation sequencing platforms — Brief Bioinform". Retrieved 2011-02-24.
- ↑ Murphy, K.; Berg, K.; Eshleman, J. (2005). "Sequencing of genomic DNA by combined amplification and cycle sequencing reaction". Clinical chemistry 51 (1): 35–39.
- ↑ Sengupta, D.; Cookson, B. (2010). "SeqSharp: A general approach for improving cycle-sequencing that facilitates a robust one-step combined amplification and sequencing method". The Journal of molecular diagnostics : JMD 12 (3): 272–277.
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407
- ↑ http://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=422803407