An Introduction to Molecular Biology/RNA:The ribonucleic acid
Ribonucleic acid is popularly known as RNA. RNA is one of the three major macromolecules (along with DNA and proteins) that are essential for all known forms of life. The chemical structure of RNA is very similar to that of DNA, with two differences--(a) RNA contains the sugar ribose while DNA contains the slightly different sugar deoxyribose (a type of ribose that lacks one oxygen atom), and (b) RNA has the nucleobase uracil while DNA contains thymine (uracil and thymine have similar base-pairing properties). Messenger RNA (mRNA) is the RNA that carries information from DNA to the ribosome, the sites of protein synthesis (translation) in the cell. The coding sequence of the mRNA determines the amino acid sequence in the protein that is produced. Many RNAs do not code for protein however (about 97% of the transcriptional output is non-protein-coding in eukaryotes). These so-called non-coding RNAs ("ncRNA") can be encoded by their own genes (RNA genes), but can also derive from mRNA introns. The most prominent examples of non-coding RNAs are transfer RNA (tRNA) and ribosomal RNA (rRNA), both of which are involved in the process of translation.There are also non-coding RNAs involved in gene regulation, RNA processing and other roles. Certain RNAs are able to catalyse chemical reactions such as cutting and ligating other RNA molecules, and the catalysis of peptide bond formation in the ribosome;these are known as ribozymes.[1]

History of RNA
[edit | edit source]Nucleic acids were discovered in 1868 by Friedrich Miescher, who called the material 'nuclein' since it was found in the nucleus.[2] It was later discovered that prokaryotic cells, which do not have a nucleus, also contain nucleic acids. The role of RNA in protein synthesis was suspected already in 1939.[3] Severo Ochoa won the 1959 Nobel Prize in Medicine (shared with Arthur Kornberg) after he discovered an enzyme that can synthesize RNA in the laboratory.[4] Ironically, the enzyme discovered by Ochoa (polynucleotide phosphorylase) was later shown to be responsible for RNA degradation, not RNA synthesis.
The sequence of the 77 nucleotides of a yeast tRNA was found by Robert W. Holley in 1965,[5] winning Holley the 1968 Nobel Prize in Medicine (shared with Har Gobind Khorana and Marshall Nirenberg).
In 1967, Carl Woese hypothesized that RNA might be catalytic and suggested that the earliest forms of life (self-replicating molecules) could have relied on RNA both to carry genetic information and to catalyze biochemical reactions—an RNA world.[6][7]
During the early 1970s, retroviruses and reverse transcriptase were discovered, showing for the first time that enzymes could copy RNA into DNA (the opposite of the usual route for transmission of genetic information). For this work, David Baltimore, Renato Dulbecco and Howard Temin were awarded a Nobel Prize in 1975. In 1976, Walter Fiers and his team determined the first complete nucleotide sequence of an RNA virus genome, that of bacteriophage MS2.[8]
In 1977, introns and RNA splicing were discovered in both mammalian viruses and in cellular genes, resulting in a 1993 Nobel to Philip Sharp and Richard Roberts. Catalytic RNA molecules (ribozymes) were discovered in the early 1980s, leading to a 1989 Nobel award to Thomas Cech and Sidney Altman. In 1990 it was found in petunia that introduced genes can silence similar genes of the plant's own, now known to be a result of RNA interference.[9][10]
At about the same time, 22 nt long RNAs, now called microRNAs, were found to have a role in the development of C. elegans.[11] Studies on RNA interference gleaned a Nobel Prize for Andrew Fire and Craig Mello in 2006, and another Nobel was awarded for studies on transcription of RNA to Roger Kornberg in the same year. The discovery of gene regulatory RNAs has led to attempts to develop drugs made of RNA, such as siRNA, to silence genes.[12]
Types of RNA
[edit | edit source]RNAs involved in protein synthesis
[edit | edit source]Messenger RNA
[edit | edit source]Messenger RNA (mRNA) is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein. In mRNA as in DNA, genetic information is encoded in the sequence of nucleotides arranged into codons consisting of three bases each. Each codon encodes for a specific amino acid, except the stop codons that terminate protein synthesis. This process requires two other types of RNA: transfer RNA (tRNA) mediates recognition of the codon and provides the corresponding amino acid, while ribosomal RNA (rRNA) is the central component of the ribosome's protein manufacturing machinery.

Ribosomal RNA
[edit | edit source]Ribosomal ribonucleic acid (rRNA) is the RNA component of the ribosome, the organelle that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity. The tRNAs bring the necessary amino acids corresponding to the appropriate mRNA codon.
-
rRNA
Transfer RNA
[edit | edit source]Transfer RNA (tRNA) is a small RNA molecule (usually about 73-95 nucleotides) that transfers a specific active amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation. It has a 3' terminal site for amino acid attachment. This covalent linkage is catalyzed by an aminoacyl tRNA synthetase. It also contains a three base region called the anticodon that can base pair to the corresponding three base codon region on mRNA. Each type of tRNA molecule can be attached to only one type of amino acid, but because the genetic code contains multiple codons that specify the same amino acid, tRNA molecules bearing different anticodons may also carry the same amino acid.
-
Tertiary structure of tRNA. CCA tail in yellow, Acceptor stem in purple, Variable loop in orange, D arm in red, Anticodon arm in blue with Anticodon in black, T arm in green.
-
The interaction of tRNA and mRNA in protein synthesis.
RNAs involved in post-transcriptional
[edit | edit source]Small nuclear ribonucleic acid (snRNA)
[edit | edit source]Small nuclear ribonucleic acid (snRNA) is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing (removal of introns from hnRNA), regulation of transcription factors (7SK RNA) or RNA polymerase II (B2 RNA), and maintaining the telomeres. They are always associated with specific proteins, and the complexes are referred to as small nuclear ribonucleoproteins (snRNP) or sometimes as snurps. These elements are rich in uridine content.
Small nucleolar RNAs (snoRNAs)
[edit | edit source]Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs which are associated with methylation, and the H/ACA box snoRNAs which are associated with pseudouridylation. snoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes.
After transcription, nascent rRNA molecules (termed pre-rRNA) are required to undergo a series of processing steps in order to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs. Methylation is the attachment or substitution of a methyl group onto various substrates. The rRNA of humans contain approximately 115 methyl group modifications. The majority of these are 2'O-ribose-methylations ( where the methyl group is attached to the ribose group). Pseudouridylation is the conversion (isomerisation) of the nucleoside uridine to a different isomeric form pseudouridine(Ψ). Mature human rRNAs contain approximately 95 Ψ modifications. Each snoRNA molecule acts as a guide for only one (or two) individual modifications in a target RNA. In order to carry out modification, each snoRNA associates with at least four protein molecules in an RNA/protein complex referred to as a small nucleolar ribonucleoprotein (snoRNP). The proteins associated with each RNA depend on the type of snoRNA molecule (see snoRNA guide families below). The snoRNA molecule contains an antisense element (a stretch of 10-20 nucleotides) which are base complementary to the sequence surrounding the base (nucleotide) targeted for modification in the pre-RNA molecule. This enables the snoRNP to recognise and bind to the target RNA. Once the snoRNP has bound to the target site the associated proteins are in the correct physical location to catalyse the chemical modification of the target base.[13]
Ribonuclease P (RNase P)
[edit | edit source]Ribonuclease P (RNase P) is a type of Ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules
-
Predicted secondary structure and sequence conservation of RNaseP_bact_a
Telomerase RNA
[edit | edit source]Telomerase RNA component, also known as TERC, is an RNA gene found in eukaryotes, that is a component of telomerase used to extend telomeres. Telomerase RNAs differ greatly in sequence and structure between vertebrates, ciliates and yeasts, but they share a 5' pseudoknot structure close to the template sequence. The vertebrate telomerase RNAs have a 3' H/ACA snoRNA-like domain.
-
3D representation of part of the telomerase RNA component. This is the solution structure of the P2b-P3 pseudoknot from human telomerase RNA
Regulatory RNAs
[edit | edit source]Antisense RNA
[edit | edit source]Antisense RNA is a single-stranded RNA that is complementary to a messenger RNA (mRNA) strand transcribed within a cell. Antisense RNA may be introduced into a cell to inhibit translation of a complementary mRNA by base pairing to it and physically obstructing the translation machinery. This effect is therefore stoichiometric. An example of naturally occurring mRNA antisense mechanism is the hok/sok system of the E.coli R1 plasmid. Antisense RNA has long been thought of as a promising technique for disease therapy; the only such case to have reached the market is the drug fomivirsen. One commentator has characterized antisense RNA as one of "dozens of technologies that are gorgeous in concept, but exasperating in ". Generally, antisense RNA still lack effective design, biological activity, and efficient route of administration.[14]
MicroRNAs (miRNAs)
[edit | edit source]MicroRNAs (miRNAs) are short ribonucleic acid (RNA) molecules, on average only 22 nucleotides long and are found in all eukaryotic cells. miRNAs are post-transcriptional regulators that bind to complementary sequences on target messenger RNA transcripts (mRNAs), usually resulting in translational repression and gene silencing. The human genome may encode over 1000 miRNAs, which may target about 60% of mammalian genes and are abundant in many human cell types.
-
The stem-loop secondary structure of a pre-microRNA from Brassica oleracea.
Small interfering RNA (siRNA)
[edit | edit source]Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA molecules, 20-25 nucleotides in length, that play a variety of roles in biology. Most notably, siRNA is involved in the RNA interference (RNAi) pathway, where it interferes with the expression of a specific gene. In addition to their role in the RNAi pathway, siRNAs also act in RNAi-related pathways, e.g., as an antiviral mechanism or in shaping the chromatin structure of a genome; the complexity of these pathways is only now being elucidated.
siRNAs were first discovered by David Baulcombe's group at the Sainsbury Laboratory in Norwich, England, as part of post-transcriptional gene silencing (PTGS) in plants.
IDENTIFICATION OF SMALL-INTERFERING RNA
By using mapping studies, it showed that target transcript cleavage corresponded to regions that they are complementary to dsRNA and occurred at 21–22-nt intervals which looks like the same size of dsRNA-derived small RNAs. To test if small RNAs mediate RISC activity, 21– 22-nt RNA duplexes with symmetric 2-nt 3′ overhangs were synthesized to mimic dsRNA- processing products. Certainly, these synthetic oligonucleotides induced target mRNA cleavage at sites corresponding to the middle of small RNA. Therefore, these dsRNA-derived small RNAs were designated as small-interfering RNAs (siRNAs). Nevertheless, a most important contribution was made by demonstration of target transcript silencing through transfection of synthetic siRNAs into mammalian cells. This work provided the foundation for numerous RNAi-based applications, including powerful "shut down" function strategy and a new potential therapeutics. As such, these milestone studies provided an spectacular example of the importance of basic science, including traditional biochemistry, in generating new areas for biomedical applications.
Structure of RNA
[edit | edit source]

Each nucleotide in RNA contains a ribose sugar, with carbons numbered 1' through 5'. A base is attached to the 1' position, in general, adenine (A), cytosine (C), guanine (G), or uracil (U). Adenine and guanine are purines, cytosine, and uracil are pyrimidines. A phosphate group is attached to the 3' position of one ribose and the 5' position of the next. The phosphate groups have a negative charge each at physiological pH, making RNA a charged molecule (polyanion). The bases may form hydrogen bonds between cytosine and guanine, between adenine and uracil and between guanine and uracil.However, other interactions are possible, such as a group of adenine bases binding to each other in a bulge, or the GNRA tetraloop that has a guanine–adenine base-pair.
An important structural feature of RNA that distinguishes it from DNA is the presence of a hydroxyl group at the 2' position of the ribose sugar. The presence of this functional group causes the helix to adopt the A-form geometry rather than the B-form most commonly observed in DNA. This results in a very deep and narrow major groove and a shallow and wide minor groove. A second consequence of the presence of the 2'-hydroxyl group is that in conformationally flexible regions of an RNA molecule (that is, not involved in formation of a double helix), it can chemically attack the adjacent phosphodiester bond to cleave the backbone.
Ribose is an aldopentose, that is a monosaccharide containing five carbon atoms that, in its open chain form, has an aldehyde functional group at one end. In the conventional numbering scheme for monosaccharides, the carbon atoms are numbered from C1' (in the aldehyde group) to C5'. The deoxyribose derivative, found in DNA, differs from ribose by having a hydrogen atom in place of the hydroxyl group in carbon C2'. Like many monosaccharides, ribose occurs in water as the linear form H-(C=O)-(CHOH)4-H and any of two ring forms: ribofuranose ("C3'-endo"), with a five-membered ring, and ribopyranose ("C2'-endo"), with a six-membered ring. The ribofuranose form is predominant in aqueous solution. The "D-" in the name D-ribose refers to the stereochemistry of the chiral carbon atom farthest away from the aldehyde group (C4'). In D-ribose, as in all D-sugars, this carbon atom has the same configuration as in D-glyceraldehyde. Ribose comprises the backbone of RNA, a biopolymer that is the basis of genetic transcription. It is related to deoxyribose, as found in DNA. Once phosphorylated, ribose can become a subunit of ATP, NADH, and several other compounds that are critical to metabolism.[15]
RNA base
[edit | edit source]adenine (A)
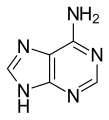
Adenine (A,) is a nucleobase (a purine derivative) with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate (ATP) and the cofactors nicotinamide adenine dinucleotide (NAD) and flavin adenine dinucleotide (FAD), and protein synthesis, as a chemical component of DNA and RNA. The shape of adenine is complementary to either thymine in DNA or uracil in RNA.
-
Adenine
cytosine (C)
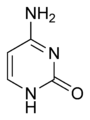
Cytosine (C) is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an amine group at position 4 and a keto group at position 2). The nucleoside of cytosine is cytidine.
-
Cytosine
guanine (G)
Guanine (G) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with conjugated double bonds.
-
Guanine
uracil (U)
-
Uracil

Found in RNA, it base-pairs with adenine and replaces thymine during DNA transcription. Methylation of uracil produces thymine.It turns into thymine to protect the DNA and to improve the efficiency of DNA replication. Uracil can base-pair with any of the bases, depending on how the molecule arranges itself on the helix, but readily pairs with adenine because the methyl group is repelled into a fixed position.[16] Uracil pairs with adenine through hydrogen bonding. Uracil is the hydrogen bond acceptor and can form two hydrogen bonds. Uracil can also bind with a ribose sugar to form the ribonucleoside uridine. When a phosphate attaches to uridine, uridine 5'-monophosphate is produced.[17]
Uracil undergoes amide-imidic acid tautomeric shifts because any nuclear instability the molecule may have from the lack of formal aromaticity is compensated by the cyclic-amidic stability. The amide tautomer is referred to as the lactam structure, while the imidic acid tautomer is referred to as the lactim structure. These tautomeric forms are predominant at pH 7. The lactam structure is the most common form of uracil.
Uracil also recycles itself to form nucleotides by undergoing a series of phosphoribosyltransferase reactions. Degradation of uracil produces the substrates aspartate, carbon dioxide, and ammonia.
- C4H4N2O2 → H3NCH2CH2COO- + NH4+ + CO2
Oxidative degradation of uracil produces urea and maleic acid in the presence of H2O2 and Fe2+ or in the presence of diatomic oxygen and Fe2+.
Uracil is a weak acid; the first site of ionization of uracil is not known.[18] The negative charge is placed on the oxygen anion and produces a pKa]] of less than or equal to 12. The basic pKa = -3.4, while the acidic pKa = 9.389. In the gas phase, uracil has 4 sites that are more acidic than water.[19]
Uracil is a common and naturally occurring pyrimidine derivative. Originally discovered in 1900, it was isolated by hydrolysis of yeast nuclein that was found in bovine thymus and spleen, herring sperm, and wheat germ. It is a planar, unsaturated compound that has the ability to absorb light.
DNA vs RNA
[edit | edit source]
RNA and DNA are both nucleic acids, but differ in three main ways.
First, unlike DNA, which is, in general, double-stranded, RNA is a single-stranded molecule in many of its biological roles and has a much shorter chain of nucleotides.
Second, while DNA contains deoxyribose, RNA contains ribose (in deoxyribose there is no hydroxyl group attached to the pentose ring in the 2' position). These hydroxyl groups make RNA less stable than DNA because it is more prone to hydrolysis.
Third, the complementary base to adenine is not thymine, as it is in DNA, but rather uracil, which is an unmethylated form of thymine.
Fourth, DNA is generally stable in alkaline conditions while RNA is unstable during alkaline condition.
Like DNA, most biologically active RNAs, including mRNA, tRNA, rRNA, snRNAs, and other non-coding RNAs, contain self-complementary sequences that allow parts of the RNA to fold and pair with itself to form double helices. Structural analysis of these RNAs has revealed that they are highly structured. Unlike DNA, their structures do not consist of long double helices but rather collections of short helices packed together into structures akin to proteins. In this fashion, RNAs can achieve chemical catalysis, like enzymes. For instance, determination of the structure of the ribosome—an enzyme that catalyzes peptide bond formation—revealed that its active site is composed entirely of RNA.
snRNA and snRNPs
[edit | edit source]snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. The action of snRNPs is essential to the removal of introns from pre-mRNA, a critical aspect of post-transcriptional modification of RNA, occurring only in the nucleus of eukaryotic cells. The two essential components of snRNPs are protein molecules and RNA. The RNA found within each snRNP particle is known as small nuclear RNA, or snRNA, and is usually about 150 nucleotides in length. The snRNA component of the snRNP gives specificity to individual introns by "recognizing" the sequences of critical splicing signals at the 5' and 3' ends and branch site of introns. The snRNA in snRNPs is similar to ribosomal RNA in that it directly incorporates both an enzymatic and a structural role. SnRNPs were discovered by Michael R. Lerner and Joan A. Steitz. Thomas R. Cech and Sidney Altman also played a role in the discovery, winning the Nobel Prize for Chemistry in 1989 for their independent discoveries that RNA can act as a catalyst in cell development.
At least five different kinds of snRNPs join the spliceosome to participate in splicing. They can be visualized by gel electrophoresis and are known individually as: U1, U2, U4, U5, and U6. Their snRNA components are known, respectively, as: U1 snRNA, U2 snRNA, U4 snRNA, U5 snRNA, and U6 snRNA. In the mid-1990s, it was discovered that a variant class of snRNPs exists to help in the splicing of a class of introns found only in metazoans, with highly-conserved 5' splice sites and branch sites. This variant class of snRNPs includes: U11 snRNA, U12 snRNA, U4atac snRNA, and U6atac snRNA. While different, they perform the same functions as do U1, U2, U4, and U6, respectively.[20]
The completed core snRNP-snurportin 1 complex is transported into the nucleus via the protein importin β. Inside the nucleus, the core snRNPs appear in the Cajal bodies, where final assembly of the snRNPs take place. This consists of additional proteins and other modifications specific to the particular snRNP (U1, U2, U4, U5). The biogenesis of the U6 snRNP occurs in the nucleus although large amounts of free U6 are found in the cytoplasm. The LSm ring may assemble first, and then associate with the U6 snRNA.
Small nuclear ribonucleic acid (snRNA) is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing (removal of introns from hnRNA), regulation of transcription factors (7SK RNA) or RNA polymerase II (B2 RNA), and maintaining the telomeres. They are always associated with specific proteins, and the complexes are referred to as small nuclear ribonucleoproteins (snRNP) or sometimes as snurps. These elements are rich in uridine content. A large group of snRNAs are known as small nucleolar RNAs (snoRNAs). These are small RNA molecules that play an essential role in RNA biogenesis and guide chemical modifications of ribosomal RNAs (rRNAs) and other RNA genes (tRNA and snRNAs). They are located in the nucleolus and the Cajal bodies of eukaryotic cells (the major sites of RNA synthesis).
RNA as an enzyme
[edit | edit source]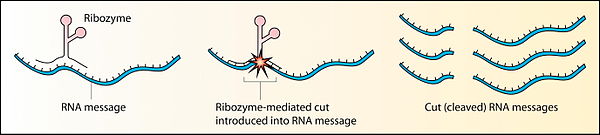
Before the discovery of ribozymes, enzymes, which are defined as catalytic proteins,[21] were the only known biological catalysts. In 1967, Carl Woese, Francis Crick, and Leslie Orgel were the first to suggest that RNA could act as a catalyst. This idea was based upon the discovery that RNA can form complex secondary structures.[22] The first ribozymes were discovered in the 1980s by Thomas R. Cech, who was studying RNA splicing in the ciliated protozoan Tetrahymena thermophila and Sidney Altman, who was working on the bacterial RNase P complex. These ribozymes were found in the intron of an RNA transcript, which removed itself from the transcript, as well as in the RNA component of the RNase P complex, which is involved in the maturation of pre-tRNAs. In 1989, Thomas R. Cech and Sidney Altman won the Nobel Prize in Chemistry for their "discovery of catalytic properties of RNA."[23] The term ribozyme was first introduced by Kelly Kruger et al. in 1982 in a paper published in the journal Cell.[24]
RNA enzymes, or ribozymes, are still found in today's DNA-based life and could be examples of living fossils. Ribozymes play vital roles, such as those in the ribosome, which is vital for protein synthesis. Many other ribozyme functions exist, for example: the Hammerhead ribozyme performs self-cleavage[25] and an RNA polymerase ribozyme can autocatalyse its own synthesis.[26]
Among the enzymatic properties important for the beginning of life are:
- The ability to self-duplicate, or duplicate other RNA molecules. Relatively short RNA molecules that can duplicate others have been artificially produced in the lab. The shortest was 165-base long, though it has been estimated that only part of the bases were crucial for this function. One version, 189-base long, had fidelity of 98.9%,[27] which would mean it would make an exact copy of an RNA molecule as long as itself in one of every eight copies. This 189 base pair ribozyme could polymerize a template of at most 14 nucleotides in length, which is too short for replication, but a promising lead for further investigation. The longest primer extension by a ribozyme polymerase was 20 bases.[28]
- The ability to catalyze simple chemical reactions which would enhance the creation of molecules which are building blocks of RNA molecules—i.e., a strand of RNA which would make creating more strands of RNA easier. Relatively short RNA molecules with such abilities have been artificially formed in the lab.[29][30]
- The ability to catalyse the formation of peptide bonds, in order to produce short peptides, or—eventually—full proteins. This is done in modern cells by ribosomes, a complex of two large RNA molecules known as rRNA and many proteins. The two rRNA molecules are thought to be responsible for its enzymatic activity. A much shorter RNA molecule has been formed in lab with the ability to form peptide bonds, and it has been suggested that rRNA has evolved from a similar molecule.[31] It has also been suggested that amino acids may have initially been complexed with RNA molecules as cofactors enhancing or diversifying their enzymatic capabilities, before evolving to the more complex peptides. mRNA may have evolved from such RNA molecules, and tRNA from RNA molecules which had catalyzed amino acid transfer to them.[32]
Although most ribozymes are quite rare in the cell, their roles are sometimes essential to life. For example, the functional part of the ribosome, the molecular machine that translates RNA into proteins, is fundamentally a ribozyme, composed of RNA tertiary structural motifs that are often coordinated to metal ions such as Mg2+ as cofactors. There is no requirement for divalent cations in a five-nucleotide RNA that can catalyze trans-phenylalanation of a four-nucleotide substrate which has three base complementary sequence with the catalyst. The catalyst and substrate were devised by truncation of the C3 ribozyme.
Ribonuclease
[edit | edit source]

Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes.
Endoribonuclease A Endoribonuclease is a ribonuclease endonuclease. It cleaves either single or double stranded RNA depending on the enzyme. Example includes both single proteins like RNase III, RNase A, RNase T1 and RNase H but also, complexes of proteins like RNase P and the RNA-induced silencing complex.
Exoribonuclease An exoribonuclease is an exonuclease ribonuclease, which are enzymes that degrade RNA by removing terminal nucleotides from either the 5' end or 3' end of the RNA molecule. Enzymes that remove nucleotides from the 5' end are called 5'-3' exoribonucleases and enzymes that remove nucleotides from the 3' end are called 3'-5' exoribonucleases.
Exoribonucleases can use either water to cleave the nucleotide-nucleotide bond (which is called hydrolytic activity) or inorganic phosphate (which is called phosphorolytic activity). Hydrolytic exoribonucleases are classified under EC number 3.1 and phosphorolytic exoribonucleases under EC number 2.7.7. As the phosphorolytic enzymes use inorganic phosphate to cleave bonds they release nucleotide disphosphates), whereas the hydrolytic enzymes (which use water) release nucleotide monosphosphates). Exoribonucleases exist in all kingdoms of life, the bacteria, archaea and eukaryotes. Exoribonucleases are involved in the degradation of many different RNA species, including messenger RNA, transfer RNA, ribosomal RNA and miRNA. Exoribonucleases can be single proteins (like RNase D or RNase PH) but also can be complexes of multiple proteins, like the exosome complex (in which four of the major exoribonuclease families are represented)[33]
RNase A RNase A is a relatively small protein (124 residues, ~13.7 kDa). It can be characterized as a two-layer α + β protein that is folded in half to resemble a taco, with a deep cleft for binding the RNA substrate. The first layer is composed of three alpha helices (residues 3-13, 24-34 and 50-60) from the N-terminal half of the protein. The second layer consist of three β-hairpins (residues 61-74, 79-104 and 105-124 from the C-terminal half) arranged in two β-sheets. The hairpins 61-74 and 105-124 form a four-stranded, antiparallel β-sheet that lies on helix 3 (residues 50-60). The longest β-hairpin 79-104 mates with a short β-strand (residues 42-45) to form a three-stranded, antiparallel β-sheet that lies on helix 2 (residues 24-34). RNase A has four disulfide bonds in its native state: Cys26-Cys84, Cys58-110, Cys40-95 and Cys65-72. The first two (26-84 and 58-110) are essential for conformational folding; each joins an alpha helix of the first layer to a beta sheet of the second layer, forming a small hydrophobic core in its vicinity. The latter two disulfide bonds (40-95 and 65-72) are less essential for folding; either one can be reduced (but not both) without affecting the native structure under physiological conditions. These disulfide bonds connect loop segments and are relatively exposed to solvent. Interestingly, the 65-72 disulfide bond has an extraordinarily high propensity to form, significantly more than would be expected from its loop entropy, both as a peptide and in the full-length protein. This suggests that the 61-74 β-hairpin has a high propensity to fold conformationally. RNase A is a basic protein (pI =8.63); its many positive charges are consistent with its binding to RNA (a poly-anion). More generally, RNase A is unusually polar or, rather, unusually lacking in hydrophobic groups, especially aliphatic ones. This may account for its need of four disulfide bonds to stabilize its structure. The low hydrophobic content may also serve to reduce the physical repulsion between highly charged groups (its own and those of its substrate RNA) and regions of low dielectric constant (the nonpolar residues). The N-terminal α-helix of RNase A (residues 3-13) is connected to the rest of RNase A by a flexible linker (residues 16-23). As shown by F. M. Richards, this linker may be cleaved by subtilisin between residues 20 and 21 without causing the N-terminal helix to dissociate from the rest of RNase A. The peptide-protein complex is called RNase S, the peptide (residues 1-20) is called the S-peptide and the remainder (residues 21-124) is called the S-protein. The dissociation constant of the S-peptide for the S-protein is roughly 30 pM; this tight binding can be exploited for protein purification by attaching the S-peptide to the protein of interest and passing a mixture over an affinity column with bound S-protein. [A smaller C-peptide (residues 1-13) also works.] The RNase S model system has also been used for studying protein folding by coupling folding and association. The S-peptide was the first peptide from a native protein shown to have (flickering) secondary structure in isolation (by Klee and Brown in 1967).[34]
RNase H In a molecular biology laboratory, as RNase H specifically degrades the RNA in RNA:DNA hybrids and will not degrade DNA or unhybridized RNA, it is commonly used to destroy the RNA template after first-strand complementary DNA (cDNA) synthesis by reverse transcription, as well as procedures such as nuclease protection assays. RNase H can also be used to degrade specific RNA strands when the cDNA oligo is hybridized, such as the removal of the poly(A) tail from mRNA hybridized to oligo(dT), or the destruction of a chosen non-coding RNA inside or outside the living cell. To terminate the reaction, a chelator, such as EDTA, is often added to sequester the required metal ions in the reaction mixture.The enzyme RNase H is a non-specific endonuclease and catalyzes the cleavage of RNA via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from archaea and prokaryota and eukaryota.
The 3-D structure of RNase H commonly consists of a 5-stranded β-sheet surrounded by a distribution of α-helices. In some RNase H, such as the one found in HIV-1, the enzyme is missing one of the helices known as the C-helix, a positively charged α-helix whose protrusive shape increases substrate binding capacity. The active site of the enzyme is centered around a conserved DEDD motif (composed of residues: D443, E478, D498, and D549) which performs the hydrolysis of the RNA substrate. A magnesium ion is commonly used as a cofactor during the hydrolysis step. It is also a potential but unconfirmed mechanism in which multiple ions are necessary for to perform the hydrolysis. The enzyme also contains a nucleic acid binding cleft about 60 Å in length that can encompass a region of 18 bound RNA/DNA base pairs. RNase H’s ribonuclease activity cleaves the 3’-O-P bond of RNA in a DNA/RNA duplex to produce 3’-hydroxyl and 5‘-phosphate terminated products. In DNA replication, RNase H is responsible for removing the RNA primer, allowing completion of the newly synthesized DNA.[35]
RNAi
[edit | edit source]
The discovery of RNAi was preceded first by observations of transcriptional inhibition by antisense RNA expressed in transgenic plants,[37] and more directly by reports of unexpected outcomes in experiments performed by plant scientists in the United States and the Netherlands in the early 1990s.[38] In an attempt to alter flower colors in petunias, researchers introduced additional copies of a gene encoding chalcone synthase, a key enzyme for flower pigmentation into petunia plants of normally pink or violet flower color. The overexpressed gene was expected to result in darker flowers, but instead produced less pigmented, fully or partially white flowers, indicating that the activity of chalcone synthase had been substantially decreased; in fact, both the endogenous genes and the transgenes were downregulated in the white flowers. Soon after, a related event termed quelling was noted in the fungus Neurospora crassa,[39] although it was not immediately recognized as related. Further investigation of the phenomenon in plants indicated that the downregulation was due to post-transcriptional inhibition of gene expression via an increased rate of mRNA degradation.[40] This phenomenon was called co-suppression of gene expression, but the molecular mechanism remained unknown.[41]

Not long after, plant virologists working on improving plant resistance to viral diseases observed a similar unexpected phenomenon. While it was known that plants expressing virus-specific proteins showed enhanced tolerance or resistance to viral infection, it was not expected that plants carrying only short, non-coding regions of viral RNA sequences would show similar levels of protection. Researchers believed that viral RNA produced by transgenes could also inhibit viral replication.[43] The reverse experiment, in which short sequences of plant genes were introduced into viruses, showed that the targeted gene was suppressed in an infected plant. This phenomenon was labeled "virus-induced gene silencing" (VIGS), and the set of such phenomena were collectively called post transcriptional gene silencing.[44]
After these initial observations in plants, many laboratories around the world searched for the occurrence of this phenomenon in other organisms.[45][46] Craig C. Mello and Andrew Fire's 1998 Nature paper reported a potent gene silencing effect after injecting double stranded RNA into Caenorhabditis elegans. In investigating the regulation of muscle protein production, they observed that neither mRNA nor antisense RNA injections had an effect on protein production, but double-stranded RNA successfully silenced the targeted gene. As a result of this work, they coined the term RNAi. Fire and Mello's discovery was particularly notable because it represented the first identification of the causative agent for the phenomenon. Fire and Mello were awarded the Nobel Prize in Physiology or Medicine in 2006 for their work.
RNAi is an RNA-dependent gene silencing process that is controlled by the RNA-induced silencing complex (RISC) and is initiated by short double-stranded RNA molecules in a cell's cytoplasm, where they interact with the catalytic RISC component argonaute. When the dsRNA is exogenous (coming from infection by a virus with an RNA genome or laboratory manipulations), the RNA is imported directly into the cytoplasm and cleaved to short fragments by the enzyme Dicer. The initiating dsRNA can also be endogenous (originating in the cell), as in pre-microRNAs expressed from RNA-coding genes in the genome. The primary transcripts from such genes are first processed to form the characteristic stem-loop structure of pre-miRNA in the nucleus, then exported to the cytoplasm to be cleaved by Dicer. Thus, the two dsRNA pathways, exogenous and endogenous, converge at the RISC complex.[47]
dsRNA cleavage
Endogenous dsRNA initiates RNAi by activating the ribonuclease protein Dicer, which binds and cleaves double-stranded RNAs (dsRNAs) to produce double-stranded fragments of 20–25 base pairs with a 2 nucleotide overhang at 3' end. Bioinformatics studies on the genomes of multiple organisms suggest this length maximizes target-gene specificity and minimizes non-specific effects.[10] These short double-stranded fragments are called small interfering RNAs (siRNAs). These siRNAs are then separated into single strands and integrated into an active RISC complex. After integration into the RISC, siRNAs base-pair to their target mRNA and induce cleavage of the mRNA, thereby preventing it from being used as a translation template.
RISC
RNA-Induced Silencing Complex, or RISC, is a multiprotein complex that incorporates one strand of a small interfering RNA (siRNA) or micro RNA (miRNA). RISC uses the siRNA or miRNA as a template for recognizing complementary mRNA. When it finds a complementary strand, it activates RNase and cleaves the RNA. This process is important both in gene regulation by microRNAs and in defense against viral infections, which often use double-stranded RNA as an infectious vector.
The active components of an RNA-induced silencing complex (RISC) are endonucleases called argonaute proteins, which cleave the target mRNA strand complementary to their bound siRNA. As the fragments produced by dicer are double-stranded, they could each in theory produce a functional siRNA. However, only one of the two strands, which is known as the guide strand, binds the argonaute protein and directs gene silencing. The other anti-guide strand or passenger strand is degraded during RISC activation.
Dicer
Dicer is an endoribonuclease in the RNase III family that cleaves double-stranded RNA (dsRNA) and pre-microRNA (miRNA) into short double-stranded RNA fragments called small interfering RNA (siRNA) about 20-25 nucleotides long, usually with a two-base overhang on the 3' end. Dicer contains two RNase III domains and one PAZ domain; the distance between these two regions of the molecule is determined by the length and angle of the connector helix and determines the length of the siRNAs it produces. Dicer catalyzes the first step in the RNA interference pathway and initiates formation of the RNA-induced silencing complex (RISC), whose catalytic component argonaute is an endonuclease capable of degrading messenger RNA (mRNA) whose sequence is complementary to that of the siRNA guide strand.[48] The human version of this gene is DICER1.
RNAi and Disease control
[edit | edit source]It may be possible to exploit RNA interference in therapy. Although it is difficult to introduce long dsRNA strands into mammalian cells due to the interferon response, the use of short interfering RNA mimics has been more successful. Among the first applications to reach clinical trials were in the treatment of macular degeneration and respiratory syncytial virus, RNAi has also been shown to be effective in the reversal of induced liver failure in mouse models.
Other proposed clinical uses center on antiviral therapies, including topical microbicide treatments that use RNAi to treat infection (at Harvard University Medical School; in mice, so far) by herpes simplex virus type 2 and the inhibition of viral gene expression in cancerous cells, knockdown of host receptors and coreceptors for HIV, the silencing of hepatitis A and hepatitis B genes, silencing of influenza gene expression, and inhibition of measles viral replication. Potential treatments for neurodegenerative diseases have also been proposed, with particular attention being paid to the polyglutamine diseases such as Huntington's disease. RNA interference is also often seen as a promising way to treat cancer by silencing genes differentially upregulated in tumor cells or genes involved in cell division. A key area of research in the use of RNAi for clinical applications is the development of a safe delivery method, which to date has involved mainly viral vector systems similar to those suggested for gene therapy.
MicroRNA (miRNA)
[edit | edit source]MicroRNAs were discovered in 1993 by Victor Ambros, Rosalind Lee and Rhonda Feinbaum during a study of the gene lin-14 in C. elegans development.They found that LIN-14 protein abundance was regulated by a short RNA product encoded by the lin-4 gene. A 61 nucleotide precursor from lin-4 gene matured to a 22 nucleotide RNA containing sequences partially complementary to multiple sequences in the 3’ UTR of the lin-14 mRNA. This complementarity was sufficient and necessary to inhibit the translation of lin-14 mRNA into LIN-14 protein. Retrospectively, the lin-4 small RNA was the first microRNA to be identified, though at the time, it was thought to be a nematode idiosyncrasy. Only in 2000 was a second RNA characterized: let-7, which repressed lin-41, lin-14, lin-28, lin-42, and daf-12 expression during developmental stage transitions in C. elegans. let-7 was soon found to be conserved in many species, indicating the existence of a wider phenomenon.
MicroRNAs (miRNAs) are short ribonucleic acid (RNA) molecules, on average only 22 nucleotides long and are found in all eukaryotic cells. miRNAs are post-transcriptional regulators that bind to complementary sequences on target messenger RNA transcripts (mRNAs), usually resulting in translational repression and gene silencing.
The function of miRNAs appears to be in gene regulation. For that purpose, a miRNA is complementary to a part of one or more messenger RNAs (mRNAs). Animal miRNAs are usually complementary to a site in the 3' UTR whereas plant miRNAs are usually complementary to coding regions of mRNAs. Perfect or near perfect base pairing with the target RNA promotes cleavage of the RNA. This is the primary mode of plant microRNAs. In animals, microRNAs more often only partially base pair and inhibit protein translation of the target mRNA (this exists in plants as well but is less common). MicroRNAs that are partially complementary to a target can also speed up deadenylation, causing mRNAs to be degraded sooner. For partially complementary microRNAs to recognise their targets, nucleotides 2–7 of the miRNA (its 'seed region') still have to be perfectly complementary.[68] miRNAs occasionally also cause histone modification and DNA methylation of promoter sites, which affects the expression of target genes. Animal microRNAs target in particular developmental genes. In contrast, genes involved in functions common to all cells, such as gene expression, have very few microRNA target sites and seem to be under selection to avoid targeting by microRNAs. dsRNA can also activate gene expression, a mechanism that has been termed "small RNA-induced gene activation" or RNAa. dsRNAs targeting gene promoters can induce potent transcriptional activation of associated genes. This was demonstrated in human cells using synthetic dsRNAs termed small activating RNAs (saRNAs), but has also been demonstrated for endogenous microRNA.[49]
miRNA and disease
Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNA been associated with disease. A manually curated, publicly available database miR2Disease documents known relationships between miRNA dysregulation and human disease.
miRNA and cancer Several miRNAs have been found to have links with some types of cancer. A study of mice altered to produce excess c-Myc — a protein with mutated forms implicated in several cancers — shows that miRNA has an effect on the development of cancer. Mice that were engineered to produce a surplus of types of miRNA found in lymphoma cells developed the disease within 50 days and died two weeks later. In contrast, mice without the surplus miRNA lived over 100 days. Leukemia can be caused by the insertion of a viral genome next to the 17-92 array of microRNAs leading to increased expression of this microRNA. Another study found that two types of miRNA inhibit the E2F1 protein, which regulates cell proliferation. miRNA appears to bind to messenger RNA before it can be translated to proteins that switch genes on and off. By measuring activity among 217 genes encoding miRNA, patterns of gene activity that can distinguish types of cancers can be discerned. miRNA signatures may enable classification of cancer. This will allow doctors to determine the original tissue type which spawned a cancer and to be able to target a treatment course based on the original tissue type. miRNA profiling has already been able to determine whether patients with chronic lymphocytic leukemia had slow growing or aggressive forms of the cancer. Transgenic mice that over-express or lack specific miRNAs have provided insight into the role of small RNAs in various malignancies. A novel miRNA-profiling based screening assay for the detection of early-stage colorectal cancer has been developed and is currently in clinical trials. Early results showed that blood plasma samples collected from patients with early, resectable (Stage II) colorectal cancer could be distinguished from those of sex-and age-matched healthy volunteers. Sufficient selectivity and specificity could be achieved using small (less than 1 mL) samples of blood. The test has potential to be a cost-effective, non-invasive way to identify at-risk patients who should undergo colonoscopy.
miRNA and heart disease The global role of miRNA function in the heart has been addressed by conditionally inhibiting miRNA maturation in the murine heart, and has revealed that miRNAs play an essential role during its development. miRNA expression profiling studies demonstrate that expression levels of specific miRNAs change in diseased human hearts, pointing to their involvement in cardiomyopathies. Furthermore, studies on specific miRNAs in animal models have identified distinct roles for miRNAs both during heart development and under pathological conditions, including the regulation of key factors important for cardiogenesis, the hypertrophic growth response, and cardiac conductance.
miRNA and the nervous system miRNAs appear to regulate the nervous system. Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (involving miR-132, miR-134 and miR-124), synapse formation and synapse maturation (where miR-134 and miR-138 are thought to be involved). Some studies find altered miRNA expression in schizophrenia.
Facts to be remembered
[edit | edit source]This is a list of RNAs in nature. Some of these categories are broad, others are single RNA families.
| Type | Abbr. | Function | Distribution | Ref. |
|---|---|---|---|---|
| Messenger RNA | mRNA | Codes for protein | All organisms | |
| Ribosomal RNA | rRNA | Translation | All organisms | |
| Signal recognition particle RNA | 7SL RNA or SRP RNA | Membrane integration | All organisms | [50] |
| Transfer RNA | tRNA | Translation | All organisms | |
| Transfer-messenger RNA | tmRNA | Rescuing stalled ribosomes | Bacteria | [51] |
| Type | Abbr. | Function | Distribution | Ref. |
|---|---|---|---|---|
| Small nuclear RNA | snRNA | Splicing and other functions | Eukaryotes and archaea | [52] |
| Small nucleolar RNA | snoRNA | Nucleotide modification of RNAs | Eukaryotes and archaea | [53] |
| SmY RNA | SmY | mRNA trans-splicing | Nematodes | [54] |
| Small Cajal body-specific RNA | scaRNA | Type of snoRNA; Nucleotide modification of RNAs | ||
| Guide RNA | gRNA | mRNA nucleotide modification | Kinetoplastid mitochondria | [55] |
| Ribonuclease P | RNase P | tRNA maturation | All organisms | [56] |
| Ribonuclease MRP | RNase MRP | rRNA maturation, DNA replication | Eukaryotes | [57] |
| Y RNA | RNA processing, DNA replication | Animals | [58] | |
| Telomerase RNA | Telomere synthesis | Most eukaryotes | [59] |
| Type | Abbr. | Function | Distribution | Ref. |
|---|---|---|---|---|
| Antisense RNA | aRNA | Transcriptional attenuation / mRNA degradation / mRNA stabilisation / Translation block | All organisms | [60][61] |
| Cis-natural antisense transcript | Gene regulation | |||
| CRISPR RNA | crRNA | Resistance to parasites, probably by targeting their DNA | Bacteria and archaea | [62] |
| Long noncoding RNA | Long ncRNA | Various | Eukaryotes | |
| MicroRNA | miRNA | Gene regulation | Most eukaryotes | [63] |
| Piwi-interacting RNA | piRNA | Transposon defense, maybe other functions | Most animals | [64][65] |
| Small interfering RNA | siRNA | Gene regulation | Most eukaryotes | [66] |
| Trans-acting siRNA | tasiRNA | Gene regulation | Land plants | [67] |
| Repeat associated siRNA | rasiRNA | Type of piRNA; transposon defense | Drosophila | [68] |
| 7SK RNA | 7SK | negatively regulating CDK9/cyclin T complex |
| Type | Function | Distribution | Ref. |
|---|---|---|---|
| Retrotransposon | Self-propagating | Eukaryotes and some bacteria | [69] |
| Viral genome | Information carrier | Double-stranded RNA viruses, positive-sense RNA viruses, negative-sense RNA viruses, many satellite viruses and reverse transcribing viruses | |
| Viroid | Self-propagating | Infected plants | [70] |
| Satellite RNA | Self-propagating | Infected cells |
| Type | Abbr. | Function | Distribution | Ref. |
|---|---|---|---|---|
| Vault RNA | vRNA | Expulsion of xenobiotics, maybe | [71] |
References
[edit | edit source]- ↑ RNA
- ↑ Dahm R (2005). "Friedrich Miescher and the discovery of DNA". Developmental Biology. 278 (2): 274–88. doi:10.1016/j.ydbio.2004.11.028. PMID 15680349.
- ↑ Caspersson T, Schultz J (1939). "Pentose nucleotides in the cytoplasm of growing tissues". Nature. 143: 602–3. doi:10.1038/143602c0.
- ↑ Ochoa S (1959). "Enzymatic synthesis of ribonucleic acid" (PDF). Nobel Lecture.
- ↑ Holley RW; et al. (1965). "Structure of a ribonucleic acid". Science. 147 (1664): 1462–65. doi:10.1126/science.147.3664.1462. PMID 14263761.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ↑ Siebert S (2006). "Common sequence structure properties and stable regions in RNA secondary structures" (PDF). Dissertation, Albert-Ludwigs-Universität, Freiburg im Breisgau. p. 1.
- ↑ Szathmáry E (1999). "The origin of the genetic code: amino acids as cofactors in an RNA world". Trends Genet. 15 (6): 223–9. doi:10.1016/S0168-9525(99)01730-8. PMID 10354582.
- ↑ Fiers W; et al. (1976). "Complete nucleotide-sequence of bacteriophage MS2-RNA: primary and secondary structure of replicase gene". Nature. 260 (5551): 500–7. doi:10.1038/260500a0. PMID 1264203.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ↑ Napoli C, Lemieux C, Jorgensen R (1990). "Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans". Plant Cell. 2 (4): 279–89. doi:10.1105/tpc.2.4.279. PMC 159885. PMID 12354959.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Dafny-Yelin M, Chung SM, Frankman EL, Tzfira T (2007). "pSAT RNA interference vectors: a modular series for multiple gene down-regulation in plants". Plant Physiol. 145 (4): 1272–81. doi:10.1104/pp.107.106062. PMC 2151715. PMID 17766396.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ↑ Ruvkun G (2001). "Glimpses of a tiny RNA world". Science. 294 (5543): 797–99. doi:10.1126/science.1066315. PMID 11679654.
- ↑ Fichou Y, Férec C (2006). "The potential of oligonucleotides for therapeutic applications". Trends in Biotechnology. 24 (12): 563–70. doi:10.1016/j.tibtech.2006.10.003. PMID 17045686.
- ↑ Small nucleolar RNA
- ↑ Antisense RNA
- ↑ RNA
- ↑ http://www.madsci.org
- ↑ Horton, Robert H.; et al.Principles of Biochemistry. 3rd ed. Upper Saddle River, NJ: Prentice Hall, 2002.
- ↑ Zorbach, W.W. Synthetic Procedures in Nucleic Acid Chemistry: Physical and Physicochemical Aids in Determination of Structure. Vol 2. New York: Wiley-Interscience, 1973.
- ↑ Lee, J.K.; Kurinovich, Ma. J Am Soc Mass Spectrom.13(8), 2005, 985-95.
- ↑ SnRNP
- ↑ Enzyme definition Dictionary.com Accessed 6 April 2007
- ↑ Carl Woese, The Genetic Code (New York: Harper and Row, 1967).
- ↑ The Nobel Prize in Chemistry 1989 was awarded to Thomas R. Cech and Sidney Altman "for their discovery of catalytic properties of RNA".
- ↑ Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (1982). "Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena". Cell. 31 (1): 147–57. PMID 6297745.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ↑ Forster AC, Symons RH (1987). "Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites". Cell. 49 (2): 211–220. doi:10.1016/0092-8674(87)90562-9. PMID 2436805.
- ↑ Johnston W, Unrau P, Lawrence M, Glasner M, Bartel D (2001). "RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension" (PDF). Science. 292 (5520): 1319–25. doi:10.1126/science.1060786. PMID 11358999.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ W. K. Johnston, P. J. Unrau, M. S. Lawrence, M. E. Glasner and D. P. Bartel RNA-Catalyzed RNA Polymerization: Accurate and General RNA-Templated Primer Extension. Science 292, 1319 (2001)
- ↑ Hani S. Zaher and Peter J. Unrau, Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA (2007), 13:1017-1026
- ↑ Huang, Yang, and Yarus, RNA enzymes with two small-molecule substrates. Chemistry & Biology, Vol 5, 669-678, November 1998
- ↑ Unrau, P. J. (1998). "RNA-catalysed nucleotide synthesis". Nature. 395 (6699): 260–263. doi:10.1038/26193. PMID 9751052.
{{cite journal}}: Cite has empty unknown parameter:|month=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ↑ Zhang, Biliang (1997). "Peptide bond formation by in vitro selected ribozymes". Nature. 390 (6655): 96–100. doi:10.1038/36375. PMID 9363898.
{{cite journal}}: Cite has empty unknown parameter:|month=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ↑ Szathmary, E. (1999). "The origin of the genetic code: amino acids as cofactors in an RNA world". Trends in Genetics. 15 (6): 223–229. doi:10.1016/S0168-9525(99)01730-8. PMID 10354582.
{{cite journal}}: Cite has empty unknown parameters:|month=and|coauthors=(help) - ↑ Ribonuclease
- ↑ http://en.wikipedia.org/w/index.php?title=Ribonuclease_A&oldid=417169401
- ↑ http://en.wikipedia.org/w/index.php?title=RNase_H&oldid=422163070
- ↑ Macrae I, Zhou K, Li F, Repic A, Brooks A, Cande W, Adams P, Doudna J (2006). "Structural basis for double-stranded RNA processing by dicer". Science. 311 (5758): 195–8. doi:10.1126/science.1121638. PMID 16410517.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Ecker JR, Davis RW (1986). "Inhibition of gene expression in plant cells by expression of antisense RNA". Proc Natl Acad Sci USA. 83 (15): 5372–5376. doi:10.1073/pnas.83.15.5372. PMC 386288. PMID 16593734.
- ↑ Napoli C, Lemieux C, Jorgensen R (1990). "Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans". Plant Cell. 2 (4): 279–289. doi:10.1105/tpc.2.4.279. PMC 159885. PMID 12354959.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Romano N, Macino G (1992). "Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences". Mol Microbiol. 6 (22): 3343–53. doi:10.1111/j.1365-2958.1992.tb02202.x. PMID 1484489.
- ↑ Van Blokland R, Van der Geest N, Mol JNM, Kooter JM (1994). "Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover". Plant J. 6: 861–77. doi:10.1046/j.1365-313X.1994.6060861.x/abs/.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Mol JNM, van der Krol AR (1991). Antisense nucleic acids and proteins: fundamentals and applications. M. Dekker. pp. 4, 136. ISBN 0824785169.
- ↑ Matzke MA, Matzke AJM. (2004). "Planting the Seeds of a New Paradigm". PLoS Biol. 2 (5): e133. doi:10.1371/journal.pbio.0020133. PMC 406394. PMID 15138502.
- ↑ Covey S, Al-Kaff N, Lángara A, Turner D (1997). "Plants combat infection by gene silencing". Nature. 385: 781–2. doi:10.1038/385781a0.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Ratcliff F, Harrison B, Baulcombe D (1997). "A Similarity Between Viral Defense and Gene Silencing in Plants". Science. 276: 1558–60. doi:10.1126/science.276.5318.1558. PMID 18610513.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Guo S, Kemphues K (1995). "par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed". Cell. 81 (4): 611–20. doi:10.1016/0092-8674(95)90082-9. PMID 7758115.
- ↑ Pal-Bhadra M, Bhadra U, Birchler J (1997). "Cosuppression in Drosophila: gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent". Cell. 90 (3): 479–90. doi:10.1016/S0092-8674(00)80508-5. PMID 9267028.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Bagasra O, Prilliman KR (2004). "RNA interference: the molecular immune system" (PDF). J. Mol. Histol. 35 (6): 545–53. doi:10.1007/s10735-004-2192-8. PMID 15614608.
- ↑ Jaronczyk K, Carmichael J, Hobman T (2005). "Exploring the functions of RNA interference pathway proteins: some functions are more RISCy than others?". Biochem J. 387 (Pt 3): 561–71. doi:10.1042/BJ20041822. PMC 1134985. PMID 15845026.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ MicroRNA
- ↑ Gribaldo1 S, Brochier-Armanet C (2006). "The origin and evolution of Archaea: a state of the art". Philos Trans R Soc Lond B Biol Sci. 361 (1470): 1007–22. doi:10.1098/rstb.2006.1841. PMC 1578729. PMID 16754611.
- ↑ Gillet R, Felden B (2001). "Emerging views on tmRNA-mediated protein tagging and ribosome rescue". Molecular Microbiology. 42 (4): 879–85. doi:10.1046/j.1365-2958.2001.02701.x. PMID 11737633.
- ↑ Thore S, Mayer C, Sauter C, Weeks S, Suck D (2003). "Crystal Structures of the Pyrococcus abyssi Sm Core and Its Complex with RNA". J. Biol. Chem. 278 (2): 1239–47. doi:10.1074/jbc.M207685200. PMID 12409299.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Kiss T (2001). "Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs". The EMBO Journal. 20 (14): 3617–22. doi:10.1093/emboj/20.14.3617. PMC 125535. PMID 11447102.
- ↑ Jones TA, Otto W, Marz M, Eddy SR, Stadler PF (2009). "A survey of nematode SmY RNAs". RNA Biol. 6 (1): 5–8. doi:10.4161/rna.6.1.7634. PMID 19106623.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Alfonzo JD, Thiemann O, Simpson L (1997). "The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria". Nucleic Acids Research. 25 (19): 3751–59. doi:10.1093/nar/25.19.3751. PMC 146959. PMID 9380494.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW (1999). "RNase P RNAs from some Archaea are catalytically active". Proc Natl Acad Sci USA. 96 (14): 7803–08. doi:10.1073/pnas.96.14.7803. PMC 22142. PMID 10393902.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Woodhams MD, Stadler PF, Penny D, Collins LJ (2007). "RNase MRP and the RNA processing cascade in the eukaryotic ancestor". BMC Evolutionary Biology. 7: S13. doi:10.1186/1471-2148-7-S1-S13. PMC 1796607. PMID 17288571.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Perreault J, Perreault J-P, Boire G (2007). "Ro-associated Y RNAs in metazoans: evolution and diversification". Molecular Biology and Evolution. 24 (8): 1678–89. doi:10.1093/molbev/msm084. PMID 17470436.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Lustig AJ (1999). "Crisis intervention: The role of telomerase". Proc Natl Acad Sci USA. 96 (7): 3339–41. doi:10.1073/pnas.96.7.3339. PMC 34270. PMID 10097039.
- ↑ Brantl S (2002). "Antisense-RNA regulation and RNA interference". Biochimica et Biophysica Acta. 1575 (1–3): 15–25. PMID 12020814.
- ↑ Brantl S (2007). "Regulatory mechanisms employed by cis-encoded antisense RNAs". Curr. Opin. Microbiol. 10 (2): 102–9. doi:10.1016/j.mib.2007.03.012. PMID 17387036.
- ↑ Brouns SJ, Jore MM, Lundgren M; et al. (2008). "Small CRISPR RNAs guide antiviral defense in prokaryotes". Science. 321 (5891): 960–4. doi:10.1126/science.1159689. PMID 18703739.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ↑ Lin S-L, Miller JD, Ying S-Y (2006). "Intronic microRNA (miRNA)". Journal of Biomedicine and Biotechnology. 2006 (4): 1–13. doi:10.1155/JBB/2006/26818. PMC 1559912. PMID 17057362.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Horwich MD, Li C Matranga C, Vagin V, Farley G, Wang P, Zamore PD (2007). "The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC". Current Biology. 17 (14): 1265–72. doi:10.1016/j.cub.2007.06.030. PMID 17604629.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Ghildiyal M, Zamore PD (2009). "Small silencing RNAs: an expanding universe". Nat. Rev. Genet. 10 (2): 94–108. doi:10.1038/nrg2504. PMC 2724769. PMID 19148191.
{{cite journal}}: Unknown parameter|month=ignored (help) - ↑ Ahmad K, Henikoff S (2002). "Epigenetic consequences of nucleosome dynamics". Cell. 111 (3): 281–84. doi:10.1016/S0092-8674(02)01081-4.
- ↑ Vazquez F, Vaucheret H (2004). "Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs". Mol. Cell. 2006 (16): 1–13. doi:10.1155/JBB/2006/26818. PMC 1559912. PMID 17057362.
- ↑ Desset S, Buchon N, Meignin C, Coiffet M, Vaury C (2008). "In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways". PLoS ONE. 3 (2): e1526. doi:10.1371/journal.pone.0001526. PMC 2211404. PMID 18253480.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Boeke JD (2003). "The unusual phylogenetic distribution of retrotransposons: a hypothesis". Genome Research. 13 (9): 1975–83. doi:10.1101/gr.1392003. PMID 12952870.
- ↑ Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F (2005). "Viroids and viroid-host interactions". Annual Review of Phytopathology. 43: 117–39. doi:10.1146/annurev.phyto.43.040204.140243. PMID 16078879.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Gopinath SC, Matsugami A, Katahira M, Kumar PK (2005). "Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound". Nucleic Acids Res. 33 (15): 4874–81. doi:10.1093/nar/gki809. PMC 1201340. PMID 16150923.
{{cite journal}}: CS1 maint: multiple names: authors list (link)