A-level Biology/Central Concepts/Meiosis, genetics and gene control
Meiosis
[edit | edit source]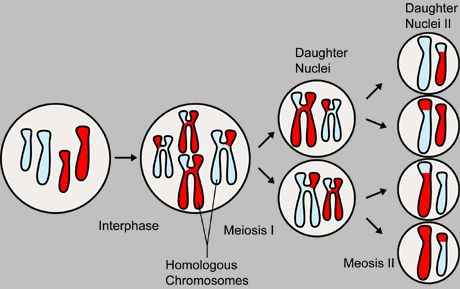
In sexual reproduction as a result of meiosis, zygotes formed from gametes, genetic variation occurs during meiosis, and genetic mutations can occur.
For A2 level, you are required to know the stages of Meiosis. https://archive.is/20130102034440/media-2.web.britannica.com/eb-media/88/78588-036-DAFF2140.jpg
Meiosis I
[edit | edit source]- Early/Middle I Prophase - Homologous chromosomes pair up, a process known as synapsis - each pair is known as a bivalent. Centrioles move to opposite ends of the nucleus as in mitosis, forming spindle fibres. Nuclear membrane begins to break down.
- Late Prophase I - Nucleolus disappears, bivalents may cross over - chromatids may break and reconnect to other chromatids - forming a chiasmata, a point where the crossing over occurs.
- Metaphase I - Bivalents line up on the spindle fibre equator, attached by centromeres.
- Anaphase I - Bivalent chromosomes move towards opposite poles within the cell. They are pulled apart because the microtubule fibres begin to shorten, pulling them along.
- Telophase I - Nucleus, nucleolus reforms. Cytokinesis also occurs - the division of cytoplasm and cell into two by constriction from edges of cell.
Meiosis II
[edit | edit source]So now we have two cells ready to divide again in meiosis II;
- Prophase II - Nuclear envelope and nucleolus disperse, centrioles replicate and move to opposite poles of the cell
- Metaphase II - Chromosomes line up separately across the equator
- Anaphase II - Chromatids (identical copies of chromosomes) separate as they are pulled towards opposite poles within the cell. They are pulled because the microtubule fibres begin to shorten, pulling them along.
- Telophase II - Nucleus, nucleolus reforms and chromosomes unravel into longer chromatin structures for reentry into interphase. Cytokinesis also occurs - the division of cytoplasm and the two cells into two by constriction from edges of cell. This forms 4 haploid cells.
Crossing over
[edit | edit source]Crossing over and independent assortment of the homologous chromosomes helps genetic variation. Crossing over is when chromatids (still in bivalent pairs) cross over, forming a chiasma. When the two formed gametes fuse at fertilisation randomly, yet more variation is produced amongst the offspring
Sections 2-6 regards genetics, the study of the way genes are passed on these chromosomes.
Alleles
[edit | edit source]A gene is a length of DNA that codes for the production of a polypeptide, and this code is held in a sequence of triplet bases. Chromosomes for making the same polypeptides are said to homologous, and a diploid cell contains one homologue from the mother and one from the father, and so in each diploid cell there are two copies of each genes, which lie in the same locus on the two homologous chromosome. The allele is a form or a variety of the same gene.
Genotype
[edit | edit source]Most genes have several alleles. We will take for example a gene that codes for eye colour (there are several, but for simplicities sake we will say there is one, and two possible colours - blue (dominant) and green (recessive))
EB - the allele for blue eyes Eg - the allele for green eyes.
Note the smaller letter for a recessive allele (g) and the bigger letter for a dominant allele (B), this is standard notation and should be used at all times.
The E stands for the locus of the gene, whilst B and g stand for blue and green. Combining these in a diploid cell might result in EB EB, EB Eg or Eg Eg. These are known as it's genotype, and there are only 3 in this case. The first and the last genotype there, are said to be homozygous since they have the same allele for that gene, and the other is heterozygous, since it has different alleles.
Phenotype
[edit | edit source]Genotype affects phenotype. If a person has EB EB, they will have blue eyes (homozygous blue), if they have EB Eg they will have blue eyes, since they have the dominant blue allele, and are so simply a carrier of the green eye allele. Only when a person has Eg Eg will they have green eyes.
Gene Inheritance
[edit | edit source]When sperm are made in the testes of a man with EB EG, a blue eyed, green carrier, two of his gametes will have the EB and two will have EG. The same with a heterozygous, blue eyed, green carrier woman. Thus, the combinations that can occur are.
The first is the man, the second is the woman;
- EB + EB = EB EB, blue eyed
- EB + Eg = EB Eg, blue eyed green carrier
- Eg + EB = EB Eg, blue eyed green carrier
- Eg + Eg = Eg Eg, green eyed
Each is individually likely, 1 in 4.
Dominance
[edit | edit source]Frequently, only one allele has an affect in a heterozygous organism, and is said to be the dominant allele to its recessive counterpart. When alleles of a gene behave like this, their symbols are written using a capital letter for the dominant allele and a lowercase letter for the recessive allele.
Typical monohybrid cross
[edit | edit source]In this example, both organisms have the Bb. They can produce s that contain either the B or b alleles. (It is conventional in to use capital letters to indicate dominant alleles and lower-case letters to indicate alleles.) The probability of an individual offspring having the genotype BB is 25%, Bb is 50%, and bb is 25%.
| Maternal | |||
|---|---|---|---|
| B | b | ||
| Paternal | B | BB | Bb |
| b | Bb | bb | |
Sex
[edit | edit source]In humans, the chromosome that determines sex is appropriately known as the sex chromosome, and the rest are autosomes. The sex chromosomes are not always homologous, and this is due to the fact there are two types of sex chromosome, the X and the Y chromosome, and the Y is shorter with fewer genes. XX = Female, XY = Male.
Linkage
[edit | edit source]If an allele for a gene is only on the X chromosome, then females will have two copies whilst men will only have one.
Dihybrid crosses
[edit | edit source]The inheritance cross of two genes at once is known as a dihybrid cross. As before, genes have two alleles, a dominant and recessive. The dominant allele is represented by a capital letter, and the recessive by a lowercase. For example, Aa. We'll use a bean plant as an example.
A = allele for purple stem a = allele for green stem
and Dd, a different locus on a chromosome, codes for leaf shape
D = allele for cut leaves d = allele for smooth leaves
So what happens when Aa/Dd (heterozygous for both), is mixed with aa/dd (homozygous for both)? When the heterozygous plant cells undergo meiosis to form gametes, the homologous chromosomes line up independently of each other on the equator during metaphase I. Usually, independent assortment occurs, with roughly half in one way, the other half lining up the other way. We can therefore predict the gametes will be of four types - AD, Ad, aD and ad, in equal numbers. The homozygous plant cells will produce gametes that are ALL ad.
This causes
- ad + AD = AaDd
- ad + Ad = Aadd
- ad + aD = aaDd
- ad + ad = aadd
This causes a 1:1:1:1 ratio of the four possible phenotypes.
Heterozygous dihybrids
[edit | edit source]If both parents are heterozygous, we have AD, Ad, aD, ad in equal proportions from both plants.
This causes:
| AD | Ad | aD | ad | |
| AD | AADD | AADd | AaDD | AaDd |
| Ad | AADd | AAdd | AaDd | Aadd |
| aD | AaDD | AaDD | aaDD | aaDd |
| ad | AaDd | Aadd | aaDd | aadd |
The result in this cross is a 9:3:3:1 phenotypic ratio, as shown by the colors, where yellow represents a round yellow (both dominant genes) phenotype, green representing a round green phenotype, red representing a wrinkled yellow phenotype, and blue representing a wrinkled green phenotype (both recessive genes).
The X2 test
[edit | edit source]The previous section shows the probability of close to 9:3:3:1 ratio of phenotypes is likely - it is unlikely that the numbers will come out exactly that. In theory, if our two plants produce 144 offspring, and were definitely heterozygous, and we have the alleles correctly we should get.
- AD = 81
- Ad = 27
- aD = 27
- ad = 9
However, we might receive results such as;
- AD = 75
- Ad = 36
- aD = 20
- ad = 16
The question is - is this close enough to be by chance or have we got something wrong? The chi-squared, or X2 test tells us just that.
The formula is below;
where
- = an observed frequency;
- = an expected (theoretical) frequency, asserted by the null hypothesis;
- = the number of possible outcomes of each event.
So, we find out the expected results, and calculate (and then square) the difference between each set of results. Divide this by the expected value, and square it to get the X2 value. This value is a probability rating for whether the difference between expected and observed results are due to chance.
So we are looking for a low probability representation in the chi-squared table [1] P = probability greater than and df = Degrees of freedom. Degrees of freedom is calculated by the amount of classes of data minus 1, since we added up all the calculated values, more data sets, larger X2 will be. A perfect result (wherein all expected = observed), would be 0, and thus basically have a probability of being to chance of 0.
Mutations
[edit | edit source]A mutation is an unpredictable change in the genetic material, and produces a different allele of a gene known as a gene mutation. Mutations can also cause changes in the structure or number of chromosomes in the cell, known as chromosome mutations. They can be caused by radiation, including UV radiation from the sun or carcinogenic chemicals. These factors are known as mutagens, and increase the chance of mutation occurring.
There are three different ways gene mutations can occur;
- Base addition - where one or more additional bases are added - CCT GAG GAG may become CCA TGA GGA G.
- Base substitution - where one based takes the place of another - CCT GAG GAG may become CCT GTG GAG.
- Base deletion - where one or more bases are deleted from the sequence - CCT GAG GAG may become CCG AGG AG.
Addition and deletion usually have a major effect on structure, and most often completely breaks the polypeptides that it codes for function. They cause frame shifts in the base sequence, causing a ripple change effect down the line for example ;
- CCT-CCT-GAG-CCG-GAG-AGG
If we remove the bolded letter during base deletion, we are left with
- CCT-CCT-AGC-CGG-AGA-GG
Which is a completely different sequence, and likely completely useless - especially since the last triplet code is not a triplet at all. However, base substitutions can sometimes have no effect, and are said to be a silent mutation - usually only if the substitution results in a STOP triplet prematurely in the sequence, and the protein synthesis will never complete.
Sickle Cell Anemia
[edit | edit source]A base substitution with significant effect is shown in sickle cell anemia where a small difference in the gene that codes for one of the amino acid chains in haemoglobin causes a big problem. When the haemoglobin is not combined with oxygen, the error causes haemoglobin to be much less soluble, causing it to stick together forming long fibres inside the red blood cells, pulling them out of shape or 'sickling' them. This makes them useless at transporting oxygen and can cause them to get stuck in capillaries.
Phenylketonuria
[edit | edit source]Another example of a base substitution causing issues is phenylketonuria, and affects an enzyme that converts phenylalanine to tyrosine (which is then converted to melanin, the brown pigment in skin and hair). If phenylalanine cannot be converted to tyrosine, then tyrosine is not converted as much as it should be to melanin, causing lighter skin and hair colour. It can also cause a serious health problem in young children, with phenylalanine accumulating in the blood and tissue fluid. Testing can prevent serious brain damage, since if the child shows positive for PKU, they will be placed on a phenylalanine-free diet.
Suggested reading: Human Genome Project (Wikipedia)
Phenotype
[edit | edit source]Your genotype affects your phenotype, but it merely gives you the potential for some things, such as height. that can be affected by your environment.
Environment
[edit | edit source]Your diet can affect how tall you are - if you have protein or vitamin deficiencies you may not grow to your full height potential, and thus your phenotype is affected. Another example is the himalayan rabbit - there is an allele that allows the formation of a dark hair pigment only at low temperatures - and thus the coldest parts of the rabbit (ears, nose, paws and tail) all receive dark hair.
Lac Operon (E.Coli)
[edit | edit source]Another example of how environment can affect phenotype is the way E.coli has genes that code for enzymes that can break down lactose (lactose permease and B galactosidase, which hydrolyse lactose to glucose and galactose). However, if the bacterium is grown in a glucose only environment, neither of the genes are switched on, but if it is moved to a lactose environment the enzymes are synthesised - said to be expressed.
The genes that regulate this are known as the lac operon (operon: a length of dna containing the base sequence that actually codes for the production of proteins, structural genes). This lac operon is show below;
The LacZ and LacY are the lactose permease and B galactosidase respectively, and ignore lacA. As you can see, before this there is a promoter. This is the section that RNA polymerase binds to catalyse the transcription of mRNA from the structural genes (lacZ, lacY). Along from the promoter is the operator, and if nothing is bound to it then the promoter is available to be bound to. What prevents this from being unbound all the time?
Elsewhere in the gene, a regulator gene codes for a repressor protein, which has two binding sites, one for the operator site and one for lactose. Usually, it is bound only to the operator site, preventing the promoter being bound to and the structural genes being expressed. However, if the repressor protein binds with lactose, it deforms the protein so that it no longer fits into the operator DNA. Thus, the promoter can bind to RNA polymerase and the structural genes are expressed.




