A-level Biology/Biology Foundation/protein structure and function
The genetic molecule must be able to carry instructions for the creation of new cells, and have the ability to be copied, exactly the same each time, many many times.
DNA & RNA Structure
[edit | edit source]DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are both macromolecules.They are also polynucleotides - polymers made from many similar molecules known as nucleotides. Both DNA and RNA can simply be known as nucleic acids. Nucleotides are joined together by condensation reactions.
Nucleotides
[edit | edit source]Nucleotides are organic compounds that consist of three joined structures: a nitrogenous base, a pentose sugar, and a phosphate group. DNA contains only four nucleotides - adenine, thymine, guanine and cytosine. RNA contains the same four except thymine is replaced with Uracil. The joined sugar is either ribose or deoxyribose, and the only difference is that deoxyribose has one fewer oxygen atoms in its molecule. There are two types of bases - purine and pyrimidine - you only really need to know that adenine and guanine are the purine bases and that purine bases are larger.
Polynucleotides
[edit | edit source]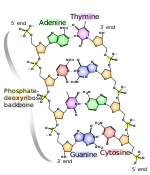
To form both DNA and RNA, many of these nucleotides are linked together in a polynucleotide chain. The structure of this poly nucleotide chain is seen in this picture: [1]. As you can see, it is formed by alternating sugars and phosphates, and the nitrogen bases project sideways. DNA molecules are simply two of these strands next to each other, running in opposite directions held together by hydrogen bonds.
In a DNA molecule, there isn't enough room for two purine strands to be opposite each other, and thus only one strand of the two can be a purine base. This however, isn't a problem since the bases are paired - complementary base pairing - Adenine always pairs with thymine, cytosine always pairs with guanine (A-T, C-G, and in RNA, it is A-U, since thymine does not appear). DNA molecules forms a 3D helix shape, bonded by hydrogen, whereas RNA remains as single strands of polynucleotide.
To the right is a rather complicated diagram, but you only really need to take note of the hydrogen bonds between the bases, the sugar phosphate backbone and the complementary base pairing - the specific structures are not required for A-Level.
DNA Replication
[edit | edit source]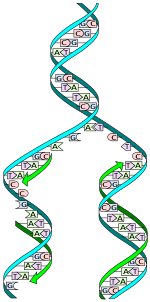
For DNA to be viable, it must have the ability to be copied perfectly many times - the discoverers of DNA, Waston and Crick suggested that the two polynucleotide strands of DNA could spilt apart, and new nucleotides (the correct, complementary ones) could line up along each strand, making a new DNA molecule. The DNA molecule can then wind back up into a double helix. This process is known as semi-conservative replication, since half of the original molecule is conserved in each of the new molecules.
Experimental proof
[edit | edit source]Nitrogen is a major constituent of DNA, and there are two different isotopes - 14N and 15N. 14N is by far the most abundant isotope of nitrogen, but DNA with the heavier 15N isotope also works.
The experiment, performed by Meselsohn and Stahn in 1958, is as follows;
E. coli, a harmless bacterium that lives in the human alimentary canal, were grown for several generations in a medium with 15N. The DNA of the resulting cells had a higher density (they were heavier). After that, E. coli cells with only 15N in their DNA were put back into a 14N medium and some were allowed to divide only once, some twice, thrice or more times. DNA was then extracted from each groups cells and was put into a solution of caesium chloride and spun in a centrifuge. The heavier the DNA ended up, the closer to the bottom of the tube it rested. The DNA that had only been grown in 15N rested nearest the bottom, whereas the group that had divided once in 14N rested in between pure 14N and pure 15N - an intermediary group, showing that that DNA had half 14N and half 15N for it's nitrogen.
DNA/RNA Synthesis
[edit | edit source]This section discusses how DNA, RNA and proteins are synthesised.
DNA Control
[edit | edit source]All cell activities are chemical reactions, and all chemical reactions are controlled by enzymes - so what controls enzymes? DNA. DNA codes for proteins, controlling which and how many proteins are made. Proteins are made from strings of amino acids, and the structure and function of a protein depends on the exact sequence of these amino acids - DNA controls this protein structure by determining the exact order in which amino acids join together.
The Triplet code
[edit | edit source]A sequence of amino acids is coded for by the sequences of nucleotides in a DNA molecule - three bases form a triplet code. Each triplet code codes for one amino acid (T-T-T, thymine-thymine-thymine stand for the amino acid lysine, for example), and it is always read in the same direction.
Genes
[edit | edit source]A gene is a part of a DNA molecule that codes for just one polypeptide, and in humans alone there is an estimated 140,000 genes. The genome is the total information in one cell, the set of genes for that organism.
Protein Synthesis
[edit | edit source]Transfer RNA (tRNA, picture below), are free molecules within the cytoplasm - in the picture at the location marked with 'a' is the anticodon, three unpaired bases, and each tRNA thus bonds with a particular amino acid. These amino acids are also free within the cytoplasm and bond at the point β in the diagram under the control of enzymes.

Process
- 1. Inside the nucleus, the DNA double helix unwinds to expose a sequence of nitrogenous bases.
- 2. Transcription - a copy of the strand is made of messenger ribonucleic acid (mRNA) which, following transcription, travels out of the nucleus through a pore in the nuclear envelope, into the main body of the cell, where protein synthesis occurs.
- 3. The mRNA couples with the small subunit of the ribosome. Transfer RNA (tRNA), brings free amino acids to the ribosome.
- 4. The anticodon present on the tRNA recognises the codon present on the mRNA string (copied from the DNA string in the nucleus), and the ribosome adds the amino acid to the growing polypeptide chain, cleaving it away from the tRNA. This process is known as translation.
- 5. As the polypeptide chain grows, it folds to form a protein.
See picture: [2]
Genetic Engineering
[edit | edit source]Since 1950, when we first determined protein synthesis as well as semi-conservative replication, we have progressed to the level where we can now change and insert DNA into cells. A good example of this is industrial insulin production.
Insulin
[edit | edit source]Diabetes mellitus is most often caused by the persons pancreas not being able to produce insulin, and victims of diabetes type I (insulin-dependent diabetes) need regular injections of insulin to survive. This insulin used to come from the pancreases of animals, such as cows or pigs - this was expensive, more risky as the insulin was not human insulin and people had ethical objections. So something had to be done - science to the rescue. The insulin gene was extracted, inserted into a bacterial vector, multiplied and extracted, details follow.
The insulin gene
[edit | edit source]To use the insulin gene (a relatively small one), it first had to be isolated - and it could not be done directly, so mRNA carrying the insulin code was extracted instead. This mRNA strand was then incubated with an enzyme called reverse transcriptase - an enzyme that reverses transcription, making DNA from RNA, two strands were made, creating a DNA double helix gene coding for insulin. To enable these insulin genes to stick onto other DNA, they were given sticky ends - a short string of guanine at each end of the polynucleotide strand.
Vector
[edit | edit source]In order to get a e. coli bacterium to replicate the insulin gene, as it is human, a vector has to be used, and in this instance the vector is known as a plasmid. A plasmid is a circular loop of DNA found in prokaryotic cells, and human DNA can be added to it. The plasmid is taken from the bacteria by dissolving their cell walls with enzymes, centrifuging them - leaving the smaller plasmids at the top. These plasmids are taken, cut with a restriction enzyme and sticky ends added - this time with cytosine ends, so that the guanine ends of the DNA strand can be attached using an enzyme known as DNA ligase. The product is called recombinant DNA.
Bacteria
[edit | edit source]The plasmids were then mixed with the bacterium E. coli - not all of them take the plasmid up, and the ones that do not are killed off with antibiotics. The ones that took the plasmid up also receive antibiotic resistance, and so only those with the plasmid survive. These genetically modified bacteria are cultured on a large scale, secreting insulin which is extracted and purified, and can then be used by people with diabetes.
Other genetic engineering
[edit | edit source]There have been many other uses of genetic engineering - we have since learnt to insert genes into any organism, and we are even beginning to have success in the field of gene therapy - replacing defective genes with good ones, which could be used to treat genetic diseases.
However, people with haemophilia have been helped massively by a protein known as human factor VIII, which several companies have produced by genetically modified hamster cells. Haemophilia is a disease in which the blood cannot clot, and this protein makes the blood clot - these people do need regular human factor VIII injections though. It is considered better than recombinant DNA from donated blood, which carries infection risks.