Structural Biochemistry/DNA Repair
Because DNA contains all of the heredity information and the instruction for protein production, it is crucial that there be very few changes to the DNA. DNA is constantly bombarded by radiation and chemical mutagens that can cause mutation. However the rate of mutation is very low because of the four main type of DNA repairs.
DNA Injury Detection and Signaling
[edit | edit source]The human genome is under constant toxic stress from normal cellular conditions such as free radicals or errors in DNA replication, as well as extrinsic conditions such as UV radiation. To combat these stresses and properly maintain the genome, the DDR pathway, or DNA damage response pathway has evolved. This pathway serves to detect errors or abnormalities, propagate the detection signal, and activate systems to correct the issue. If the damage is irreparable, the cell undergoes apoptosis, or programmed cell death, to avoid passing on the potentially lethal errors in DNA. Cells come across DNA damage constantly, so the DDR pathway is vital to cell survival.
The most lethal form of DNA damage comes from ionizing radiation which causes breaks in the double stand. The repair protein RAD51 quickly collects into foci at sites of DNA damage. It is suggested that damaged induced phosphorylation of the histone variant H2AX indicates the sites of DNA breaks; many other repair proteins also collect at these sites of H2AX accumulation. In mice lacking H2AX, immune system degradation and increased incidence of tumors are found.
The major regulators of cellular response to DNA damage are ATM and ATR kinases (ataxia telangiectasia mutated) through the regulation of phosphorylation of over 700 proteins. This phosphorylation is the initial step in the signaling of DNA damage.
“Structural Dynamics in DNA damage signaling and repair” was an article written by JJ Perry, Elizabeth Cotner-Gohara, Tom Ellenberger, and John A. Tainer. In this article, DNA damage responses are studied in aspects that reveal the role of protein in such pathways. DNA is continually damaged by metabolites and toxicants. Thus, DNA repair and damage response are essential in the function of life. There are three steps in which DNA damage is involved. The damage is first detected, removed, then eventually replaced with the correct DNA sequence. The pathway regenerates a 3’ terminal that will be extended using DNA polymerase with an undamaged strand as the template. The repair is completed with a ligase resealing the DNA backbone. Because this process of repair generates toxic intermediates, strong “genetic selection” is required as the DNA is being restored. Proteins structures are found to be connected to the coordination of steps within the DNA damage response and repair pathways. This is very important because proteins are once again, related in the DNA replication process.
When different methods come together, the dynamics of DNA repair complexes can be studied in great details. Such methods involve X-ray crystallography, NMR, SAXS – small-angle X-ray scattering, DXMS – hydrogen-deuterium exchange mass spectrometry, etc. These methods provide information as small as from the nanoscale to atomic level. For instance, SAXS gives information on the flexibility of macromolecules in solutions. It also provides information on the entire pathways and their interactions in solution. In addition, DXMS shows more on the conformation changes that take place during the repairing process as detailed as the resolution of a single amino acid. Thus, combining different structural biochemistry methods helps scientist in discovering the different coordination’s between DNA repair and damage response system. Current studies found that the “Transition between different enzyme conformations can involve non-native interactions that lower the energy barrier for inter-conversion between different states” (1). This discovery is very important because it describes the connections between the changes in the DNA repair complex (conformation changes) and the biological outcomes occurred through such changes. For instance, as stated, the changes in enzyme conformation cause the lower of activation energy for the conversion between different states during the process of restoring damaged DNA. Another example is that changing the normal protein flexibility and the stability of the repair protein system can cause great genetic diseases. Changes in DNA and ATP binding are found to be related to cancer as well as how the defects in the flexibility and stability of DNA repair framework are related to aging disorders such as Cockayne Syndrome or TTD.
The damage repair is carried out by the multi-domain nucleotide excision repair helicase (NER). This enzyme removes bulky and distorted cut from one strand of the DNA needed to be repaired. This is a very precise process where only the defected strand is removed without affecting the undamaged DNA strand because the undamaged DNA strand serves as the template for the modification and repairing process. The NER proteins are assembled in a way that allows for the verification of the damaged site before the actual removal of the DNA backbone. One example of DNA repairing process is on the performance of Yeast Rad4, a multi-domain protein that binds to the distorted part of the helix being repaired by NER. The binding of the protein is showed to stabilize the distorted DNA structure. Observations show that Rad4 inserts a beta hairpin through the DNA helix to relocate its bases. One surprising discovery was that instead of binding to the damaged DNA strand, Rad4 is bound to the undamaged one. The result was that the helical axis is offset due to the damaged DNA strand, causing a bend in structure that increase the Rad4 DNA interaction surface to the neighboring hairpin regions. This extending interaction creates a more stabilized damaged DNA, though its bases are now exposed to the solvent. This stabilization aids NER as it is repairing the damaged strand.
Another important component in the DNA damage response and repair is BER – base excision repair pathway. The difference between BER and NER is that BER has the ability to detect and remove single nucleotides with the smallest modification such as the addition of one single methyl group. Thus, it is extremely efficient in fixing distorted DNA strands. In BER, the oxidative damage-specific glycosylates OGG1 and MutM are found to interact with 8-oxoG bases. 8-oxoG bases are composed of a hydrogen-bond donor N9 and an accept O8. They interact with OGG1 and provides selective cut of the damaged DNA. This entire complex is known as the pseudo-Michaelis complex. Overall, different mechanisms were observed in the process of DNA damage response and repair from the combination of methods ranging from NMR, X-ray crystallography, to SAXS, etc.
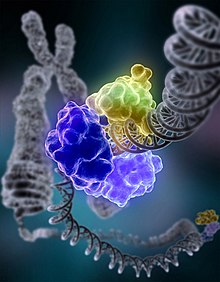
Below is an image of a process of DNA repair where the DNA ligase I is repairing a chromosomal damage.
Role of 9-1-1 in DNA Repair
[edit | edit source]DNA repair consists of the detection of existing damage and the actual healing of this impairment. 9-1-1 is a heterotrimeric protein, consists of three sub-units in which at least one is different than the other two, that wraps around DNA to initiate the recruitment of specific checkpoint proteins and freezes the cell cycle temporarily. More specifically, it causes phosphorylation of Sc-Mec1/Hs-ataxia telangiectasia, where Sc- and Hs- prefixes refer to Saccharomyces cerevisiae (a eukaryotic species) and Homo sapiens respectively, and Rad3. Chk1 and Sc-Rad53/Hs-CHK2 protein kinases are activated resulting in the inhibition of cell cycle phases G1/S intra-S or G2/M. Accumulation of repair genes, fixation of the replication fork, and the decrease in production of cyclins (proteins that progress the cell cycle) also result from this activation. 9-1-1 works with Sc-Cdc28 to selectively accumulate Sc-Ddc2. The presence of Sc-Ddc2/Hs-ATRIP, Sc-Mec1/HS-ATR, and 9-1-1 together activates the checkpoint regardless of the detection of DNA damage.
Mismatch Repair
[edit | edit source]Mismatch repairs corrects any mistakes in nucleotide pairing that escape the proofreading ability of DNA polymerase during replication. Base nucleotides that are incorrectly paired causes deformity in the secondary structure of DNA. The MSH2 and MSH6 dimer binds to the mismatch on the strand. Then, MLH1, an endonuclease, will bind to the MSH and nick the strand. Then exonucleases will degrade the region in between and then allow DNA polymerase delta to place the correct nucleotide and DNA ligase will re-connect the strand. Using this ability, the enzyme cut out the distorted portion of the new DNA strand and then use the old DNA strand as a template to fill in the gap. In E.Coli, the mismatch repair enzyme recognizes the old DNA strand by the presence of methyl groups on certain sequences. In eukaryotic cells, it is unknown how the enzyme is able to distinguish between the old and new DNA strands.
Direct Repair
[edit | edit source]
In direct repair, instead of replacing an entire nucleotide, the wrong nucleotide is structurally changed to the right nucleotide. UV ray from the sun causes pyrimidine dimers by forming covalent bonding between adjacent pyrimidines. Some eukaryotic cells have an enzyme called photolyase. The enzyme breaks the covalent bond between the pyrimidine dimers with the energy from light.
Nucleotide Excision
[edit | edit source]NER Helicase
[edit | edit source]DNA repair is carried out by the nucleotide excision repair (NER) helicase, a protein that is composed of multiple domains. NER assembles around damaged DNA regions (which, because of their error, contain a bulge or lesion that encourages NER to bind) in a stepwise manner, allowing damage to be carefully verified before the actual excision is performed. For example, yeast Rad4 protein (an analogue of mammalian XPC) indirectly detects DNA damage by binding to a nearby undamaged region. The damaged DNA strand is flexible, allowing a stable complex to form which includes Rad23, the protein that actually repairs the damage.
If XPC-Rad4 cannot detect a damaged site, one alternative involves the DDC1-DDC2 dimer. This dimer forms a complex with a damaged DNA region and an ubiquitin ligase. The complex ubiquitinates XPC and DDC2, the latter of which then releases the DNA molecule, passing it on to XPC and the normal NER process.
Nucleotide Excision Repair can be divided into two subcategories: Global Genome Repair and Transcription Coupled Repair.
Global Genome Repair involves the XPC and hHR23B dimer binding to the damages DNA and then Transcription Factor 2H (TFIIH) bind to the complex. Then XPG binds and the DNA is further unwound. The nucleases XPG and XPF cleave the DNA, which essentially removes the damaged DNA. Then DNA polymerase delta fills in the gap with the correct nucleotide and then DNA ligase re-connects the strand.
Transcription coupled repair is when RNA polymerase stalls at the damaged site and then Cockayne Syndrome B protein (CSB) displaces RNA polymerase and recruits TFIIH and XPG. The DNA is unwound before the nucleases XPG and XPF cleave the DNA. Then the damaged section is removed and DNA polymerase delta fills in the gap and ligase re-connects the strand.
Source: Molecular Cell Biology, Lodish et al., 6th edition (2008), pages 145-160
The Base-Excision Repair pathway
[edit | edit source]
Not all damages are large enough to cause the lesions that are detected by NER. The base excision repair (BER) pathway repairs single nucleotide errors, sometimes as slight as the addition of a methyl group. While small, these damages can often be enough to impede DNA replication or produce nonfunctional proteins. Damage detection in the BER pathway is difficult because, in addition to the errors being small, there are a large number of them. Numerous enzymes are used to detect different small errors and initiate the BER pathway.
The first step in base-excision repair is the excision of modified nucleotide. Enzymes called DNA glycosylases, each has its own ability to recognize certain type of modified bases, cleave the bond between the 1'-carbon of the deoxyribose sugar and the base and remove the base. Then enzyme called apurinic or apyrimidinic (AP) endonuclease breaks the phosphodiester bond and another enzyme removes the deoxyribose sugar. DNA polymerase comes and adds the correct nucleotide to a free 3'OH group. Finally, DNA ligase connects the DNA strand by forming phosphodiester bond.
Backbone repair and DNA ligase
[edit | edit source]Damage to the sugar-phosphate backbone of DNA is repaired by DNA ligases. Because the DNA backbone is common to all organisms, these ligases are likewise found in every organism that uses DNA as its genetic material. DNA ligase seals breaks in the backbone by a three-step process. In the first step, several of the enzyme's domains adopt a specific conformation, allowing an active site lysine residue to be adenylated. In the last two steps, the enzyme encircles the broken DNA strand and fuse the two ends together.
Double-Strand Break Repair
[edit | edit source]Breaks in the double strand of DNA are common, but particularly hazardous to the cell due to increased chance of genetic mutation. Major causes of double strand breaks include reactive oxygen from oxidative metabolism, ionizing radiation, and enzyme errors. The strand could be repaired in one of two major ways: homologous-directed repair and the nonhomologous DNA end joining pathway (NHEJ).
Homology-Directed Repair
[edit | edit source]Any diploid organism could use homology-directed repair, even if the diploidy is temporary, as in bacteria. Types of homology-directed repair include homologous recombination, single strand annealing, and breakage-induced replication. In homologous recombination, an identical or nearly identical sequence of DNA is required as a template for repair during the S phase of the cell cycle, which occurs only during and shortly after DNA replication, and before mitosis. Nucleotide sequences are then exchanged between similar strands.
Nonhomologous DNA End Joining Pathway (NHEJ)
[edit | edit source]NHEJ arose as an alternative to homology-directed repair, as template donors are usually not available in nondividing cells. With a remarkably flexible mechanism, NHEJ has a wide diversity of substrates that can be converted into the desired product. Like other DNA repair processes, it requires three main proteins: a nuclease to resect damaged DNA , polymerases to fill in new DNA, and a ligase to the restore the strand. Key components include Ku, DNA-PKcs, Artemis, Pol x polymerases, and the ligase complex consisting of XLF, XRCC4, and DNA ligase IV. Each DNA end could then be modified independently multiple times, and substitutions with other enzymes is permitted due to its flexible nature. The problem of joining heterogenous DNA ends at double-strand breaks was shown to have evolved convergently in prokaryotes and eukaryotes.
References
[edit | edit source]- Huen, M. SY. "Assembly of checkpoint and repair machineries at DNA damage sites." Trends in Biochemical Sciences, Volume 35, Issue 2, 101-108, 28 October 2009
- Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. “Structural dynamics in DNA damage signaling and repair.” Curr. Opin. Struct. Biol. 2010 Jun; 20(3)
- Pierce, Benjamin A., Jung H. Choi, and Mark E. McCallum. Genetics: a Conceptual Approach. New York, NY: W.H. Freeman, 2008. Print.
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211.
- Perry, J. Jefferson P., Elizabeth Cotner-Gohara, Tom Ellenberger, and John A Tainer. “Structural Dynamics in DNA Damage Signaling and Repair”. Current Opinion in Structural Biology. (2010): 283-294. ScienceDirect.
- Eichinger, S. Christian and Stefan Jentsch. "9-1-1: PCNA's specialized cousin." Trends in Biochemical Sciences, Volume 36, Issue 11, 563-568, 04 October 2011.

