Data Mining Algorithms In R/Clustering/Self-Organizing Maps (SOM)
Introduction
[edit | edit source]The Kohonen Self-Organizing Feature Map (SOFM or SOM) is a clustering and data visualization technique based on a neural network viewpoint. As with other types of centroid-based clustering, the goal of SOM is to find a set of centroids (reference or codebook vector in SOM terminology) and to assign each object in the data set to the centroid that provides the best approximation of that object. In neural network terminology, there is one neuron associated with each centroid [1].
As with incremental K-means, data objects are processed one at a time and the closest centroid is updated. Unlike K-means, SOM impose a topographic ordering on the centroids and nearby centroids are also updated. The processing of points continues until some predetermined limit is reached or the centroids are not changing very much. The final output of the SOM technique is a set of centroids that implicitly define clusters. Each cluster consist of the points closest to a particular centroid [1].
SOM is a clustering technique that enforces neighborhood relationships on the resulting cluster centroids. Because of this, clusters that are neighbors are more related to one another than clusters that are not. Such relationships facilitate the interpretation and visualization of the clustering results. Indeed, this aspect of SOM has been exploited in many areas, such as visualizing Web documents or gene array data[1].
Algorithm
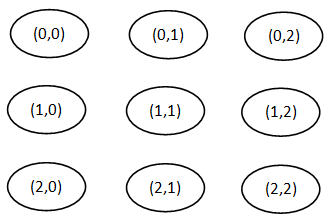
[edit | edit source]A distinguishing feature of SOM is that it imposes a topographic(spacial) organization on the centroids (neurons). Figure 1 shows an example of a two-dimensional SOM in which the centroids are represented by nodes that are organized in a rectangular lattice. Each centroid is assigned a pair of coordinates(i,j). Sometimes, such a network is drawn with links between adjacent nodes, but can be misleading because the influence of one centroid on another is via a neighborhood that is defined in terms of coordinates, not links. There are many types of SOM neural networks, but it will be focus on to two-dimensional SOMs with a rectangular or hexagonal organization of the centroids.[1]
Figure 1: Two-dimensional 3-by-3 rectangular SOM neural network [1].
Even though SOM is similar to K-means, there is a fundamental difference. Centroids used in SOM have a predetermined topographic ordering relationship. During the training process, SOM uses each data point to update the closest centroid and centroids that are nearby in the topographic ordering. In this way, SOM produces an ordered set of centroids for any given data set. In other words, the centroids that are close to each other in the SOM grid are more closely related to each other than to the centroids that are farther away. Because of this constraint, the centroids of a two-dimensional SOM can be viewed as lying on a two-dimensional surface that tries to fit the n-dimensional data as well as possible. The SOM centroids can also be thought of as the result of a nonlinear regression with respect to the data points. At a high level, clustering using the SOM technique consists of the steps described in Algorithm below[1]:
1: Initialize the centroids. 2: repeat 3: Select the next object. 4: Determine the closest centroid to the object. 5: Update this centroid and the centroids that are close, i.e., in a specified neighborhood. 6: until The centroids don't change much or a threshold is exceeded. 7: Assign each object to its closest centroid and return the centroids and clusters.
Implementation
[edit | edit source]For R (R Development Core Team 2007), three packages are available from the Comprehensive R Archive Network (CRAN) implementing standard SOMs [2]
- The kohonen package implements self-organizing maps as well as some extensions for supervised pattern recognition and data fusion.
- The som package provides functions for self-organizing maps.
- The wccsom package SOM networks for comparing patterns with peak shifts.
For this discussion the focus is on the kohonen package because it gives SOM standards features and order extensions. The R package kohonen provides functions for self-organizing maps. It also provides two extensions that allow the use of SOMs for classification and regression tasks as well as data mining tasks. It specifically emphasizes visualisation. The basic functions are: som for the usual unsupervised form of self-organizing maps; xyf for supervised self-organizing maps and X-Y fused maps, which are useful when additional information in the form of, e.g., a class variable is available for all objects; bdk, an alternative formulation called bi-directional Kohonen maps; and finally, from version 2.0.0 on, the generalisation of the xyf maps to more than two layers of information, in the function supersom. These functions can be used to define the mapping of the objects in the training set to the units of the map [3].
Several data sets are included in the kohonen package: the wine data from the UCI Machine Learning Repository[4], near-infrared spectra from ternary mixtures of ethanol, water and iso-propanol, measured at different temperatures described by Wülfert et al. (1998) [5], and finally a set of microarray data, the yeast data from Spellman et al. (1998)[6]. The wine data set contains information on a set of 177 Italian wine samples from three different grape cultivars; thirteen variables (such as concentrations of alcohol and flavonoids, but also color hue) have been measured. The yeast data are a subset of the original set containing 6178 genes, which are assumed to be related to the yeast cell cycle. The set contains 800 genes for which, using six different synchronization methods, time-dependent expressions have been measured [3].
The different types of self-organizing maps can be obtained by calling the functions som, xyf, bdk, or supersom, with the appropriate data representation as the first argument(s). Several other arguments provide additional parameters, such as the map size, the number of iterations, etcetera. The object that is returned can then be used for inspection, plotting, mapping, and prediction. Below we will show the functions available in the package. Visualization functions will be discussed at Visualization topic [3].
Function som
[edit | edit source]Function som implement the standard form of self-organizing maps [7].
som(data, grid=somgrid(), rlen = 100, alpha = c(0.05, 0.01), radius = quantile(nhbrdist, 0.67) * c(1, -1), init, toroidal = FALSE, n.hood, keep.data = TRUE)
the arguments are:
- data: a matrix, with each row representing an object.
- grid: a grid for the representatives.
- rlen: the number of times the complete data set will be presented to the network.
- alpha: learning rate, a vector of two numbers indicating the amount of change. Default is to decline linearly from 0.05 to 0.01 over rlen updates.
- radius: the radius of the neighbourhood, either given as a single number or a vector (start, stop). If it is given as a single number the radius will run from the given number to the negative value of that number; as soon as the neighbourhood gets smaller than one only the winning unit will be updated. The default is to start with a value that covers 2/3 of all unit-to-unit distances.
- init: the initial representatives, represented as a matrix. If missing, chosen (without replacement) randomly from ’data’.
- toroidal: if TRUE, the edges of the map are joined. Note that in a hexagonal toroidal map,the number of rows must be even.
- n.hood: the shape of the neighbourhood, either "circular" or "square". The latter is the default for rectangular maps, the former for hexagonal maps.
- keep.data: save data in return object.
return an object of class "kohonen" with components:
- data: data matrix, only returned if keep.data == TRUE.
- grid: the grid, an object of class "somgrid".
- codes: a matrix of code vectors.
- changes: vector of mean average deviations from code vectors.
- unit.classif: winning units for all data objects, only returned if keep.data == TRUE.
- distances: distances of objects to their corresponding winning unit, only returned if keep.data == TRUE.
- toroidal: whether a toroidal map is used.
- method: the type of som, here "som".
Function xyf
[edit | edit source]Function xyf is a supervised version of self-organizing maps for mapping high-dimensional spectra or patterns to 2D [7].
xyf(data, Y, grid=somgrid(), rlen = 100, alpha = c(0.05, 0.01), radius = quantile(nhbrdist, 0.67) * c(1, -1), xweight = 0.5, contin, toroidal = FALSE, n.hood, keep.data = TRUE)
the arguments are:
- data: a matrix, with each row representing an object.
- Y: property that is to be modelled. In case of classification, Y is a matrix of zeros, with exactly one ’1’ in each row indicating the class. For prediction of continuous properties, Y is a vector. A combination is possible, too, but one then should take care of appropriate scaling.
- grid: a grid for the representatives.
- rlen: the number of times the complete data set will be presented to the network.
- alpha: learning rate, a vector of two numbers indicating the amount of change. Default is to decline linearly from 0.05 to 0.01 over rlen updates.
- radius: the radius of the neighbourhood, either given as a single number or a vector start, stop). If it is given as a single number the radius will run from the given number to the negative value of that number; as soon as the neighbourhood gets smaller than one only the winning unit will be updated. The default is to start with a value that covers 2/3 of all unit-to-unit distances.
- xweight: the weight given to the X map in the calculation of distances for updating Y. Default is 0.5.
- contin: parameter indicating whether Y is continuous or categorical. The default is to check whether all row sums of Y equal 1: in that case contin is FALSE.
- toroidal: if TRUE, the edges of the map are joined. Note that in a hexagonal toroidal map, the number of rows must be even.
- n.hood: the shape of the neighbourhood, either "circular" or "square". The latter is the default for rectangular maps, the former for hexagonal maps.
- keep.data: save data in return value.
return an object of class "kohonen" with components:
- data: data matrix, only returned if keep.data == TRUE.
- Y: Y, only returned if keep.data == TRUE.
- contin: parameter indicating whether Y is continuous or categorical.
- grid: the grid, an object of class "somgrid".
- codes: list of two matrices, containing codebook vectors for X and Y, respectively.
- changes: matrix containing two columns of mean average deviations from code vectors. Column 1 contains deviations used for updating Y; column 2 for updating X.
- toroidal: whether a toroidal map is used.
- unit.classif: winning units for all data objects, only returned if keep.data == TRUE.
- distances: distances of objects to their corresponding winning unit, only returned if keep.data == TRUE.
- method: the type of som, here "xyf".
Function bdk
[edit | edit source]Supervised version of self-organising maps for mapping high-dimensional spectra or patterns to 2D: the Bi-Directional Kohonen map [7].
bdk(data, Y, grid=somgrid(), rlen = 100, alpha = c(0.05, 0.01), radius = quantile(nhbrdist,0.67) * c(1, -1), xweight = 0.75, contin, toroidal = FALSE, n.hood, keep.data = TRUE)
the arguments are:
- data: a matrix, with each row representing an object.
- Y: property that is to be modelled. In case of classification, Y is a matrix with exactly one ’1’ in each row indicating the class, and zeros elsewhere. For prediction of continuous properties, Y is a vector. A combination is possible, too, but one then should take care of appropriate scaling.
- grid: a grid for the representatives.
- rlen: the number of times the complete data set will be presented to the network.
- alpha: learning rate, a vector of two numbers indicating the amount of change. Default is to decline linearly from 0.05 to 0.01 over rlen updates.
- radius: the radius of the neighbourhood, either given as a single number or a vector (start, stop). If it is given as a single number the radius will run from the given number to the negative value of that number; as soon as the neighbourhood gets smaller than one only the winning unit will be updated. The default is to start with a value that covers 2/3 of all unit-to-unit distances.
- xweight: the initial weight given to the X map in the calculation of distances for updating Y, and to the Y map for updating X. This will linearly go to 0.5 during training. Defaults to 0.75.
- contin: parameter indicating whether Y is continuous or categorical. The default is to check whether all row sums of Y equal 1: in that case contin is FALSE.
- toroidal: if TRUE, the edges of the map are joined. Note that in a hexagonal toroidal map, the number of rows must be even.
- n.hood: the shape of the neighbourhood, either "circular" or "square". The latter is the default for rectangular maps, the former for hexagonal maps.
- keep.data: save data in return value.
return an object of class "kohonen" with components:
- data: data matrix, only returned if keep.data == TRUE.
- Y: Y, only returned if keep.data == TRUE.
- contin: parameter indicating whether Y is continuous or categorical.
- grid: the grid, an object of class "somgrid".
- codes: list of two matrices, containing codebook vectors for X and Y, respectively.
- changes: matrix containing two columns of mean average deviations from code vectors. Column 1 contains deviations used for updating Y; column 2 for updating X.
- toroidal: whether a toroidal map is used.
- unit.classif: winning units for all data objects, only returned if keep.data == TRUE.
- distances: distances of objects to their corresponding winning unit, only returned if keep.data== TRUE.
- method: the type of som, here "bdk".
Function supersom
[edit | edit source]An extension of xyf maps to multiple data layers, possibly with different numbers of variables (though equal numbers of objects) [7].
supersom(data, grid=somgrid(), rlen = 100, alpha = c(0.05, 0.01), radius = quantile(nhbrdist, 0.67) * c(1, -1), contin, toroidal = FALSE, n.hood, whatmap = NULL, weights = 1, maxNA.fraction = .5, keep.data = TRUE)
the arguments are:
- data: list of data matrices.
- grid: a grid for the representatives: see somgrid.
- rlen: the number of times the complete data set will be presented to the network.
- alpha: learning rate, a vector of two numbers indicating the amount of change. Default is to decline linearly from 0.05 to 0.01 over rlen updates.
- radius: the radius of the neighbourhood, either given as a single number or a vector (start, stop). If it is given as a single number the radius will run from the given number to the negative value of that number; as soon as the neighbourhood gets smaller than one only the winning unit will be updated. The default is to start with a value that covers 2/3 of all unit-to-unit distances.
- contin: parameter indicating whether data are continuous or categorical. The default is to check whether all row sums equal 1: in that case contin is FALSE.
- toroidal: if TRUE, the edges of the map are joined. Note that in a hexagonal toroidal map, the number of rows must be even.
- n.hood: the shape of the neighbourhood, either "circular" or "square". The latter is the default for rectangular maps, the former for hexagonal maps.
- whatmap: For supersom maps: what layers to use in the mapping.
- weights: the weights given to individual layers. Default is 1/n, with n the number of layers.
- maxNA.fraction: the maximal fraction of values that may be NA to prevent the row or column to be removed.
- keep.data: save data in return value.
return an object of class "kohonen" with components:
- data: data matrix, only returned if keep.data == TRUE.
- contin: parameter indicating whether elements of data are continuous or categorical.
- na.rows: indices of objects (rows) that are removed because at least one of the layers has to many NAs for these objects.
- unit.classif: winning units for all data objects, only returned if keep.data == TRUE.
- distances: distances of objects to their corresponding winning unit, only returned if keep.data == TRUE.
- grid: the grid, an object of class somgrid.
- codes: a list of matrices containing codebook vectors.
- changes: matrix of mean average deviations from code vectors; every map corresponds with one column.
- toroidal: whether a toroidal map is used.
- n.hood: the shape of the neighbourhood, either "circular" or "square". The latter is the
- default for rectangular maps, the former for hexagonal maps.
- weights: For supersom maps: weights of layers uses in the mapping.
- whatmap: For supersom maps: what layers to use in the mapping.
- method: type of map, here "supersom".
Function predict.kohonen
[edit | edit source]Map objects to a trained Kohonen map, and return for each object the property associated with the corresponding winning unit [7].
## S3 method for class 'kohonen': predict(object, newdata, trainX, trainY, unit.predictions, threshold = 0, whatmap = NULL, weights = 1, ...)
the arguments are:
- object: Trained network.
- newdata: Data matrix for which predictions are to be made. If not given, defaults to the training data (when available).
- trainX: Training data for obtaining predictions for unsupervised maps; necessary for som maps trained with the keep.data = FALSE option.
- trainY: Values for the dependent variable for the training data; necessary for som and supersom maps.
- unit.predictions: Possible override of the predictions for each unit.
- threshold: Used in class predictions; see classmat2classvec.
- whatmap: For supersom maps: what layers to use in the mapping.
- weights: For supersom maps: weights of layers uses in the mapping.
- ...: Currently not used.
returns a list with components:
- prediction: predicted values for the properties of interest. When multiple values are predicted, this element is a list, otherwise a vector or a matrix.
- unit.classif: unit numbers to which objects in the data matrix are mapped.
- unit.predictions: mean values associated with map units. Again, when multiple properties are predicted, this is a list.
Function classvec2classmat
[edit | edit source]Convert a classification vector into a matrix or the other way around [7].
classvec2classmat(yvec) classmat2classvec(ymat, threshold=0)
the arguments are:
- yvec: class vector. Usually integer values, but other types are also allowed.
- ymat: class matrix: every column corresponds to a class.
- threshold: only classify into a class if the probability is larger than this threshold.
return:
- classvec2classmat: returns the classification matrix, where each column consists of zeros and ones.
- classmat2classvec: returns a class vector (integers).
Function check.whatmap
[edit | edit source]Check the validity of a whatmap argument [7].
check.whatmap(x, whatmap)
the arguments are:
- x: Either a kohonen object from supersom, or a list of data matrices that can be used as input data for supersom.
- whatmap: An indication of a subset of the data; either by naming the elements, or giving indices. If whatmap equals NULL, no selection is performed.
Returns:
- Returns a numerical vector with the indices of the selected layers.
Function map.kohonen
[edit | edit source]Map a data matrix onto a trained SOM [7].
## S3 method for class 'kohonen': map(x, newdata, whatmap = NULL, weights, scale.distances = (nmaps > 1), ...)
the arguments are:
- x: A trained supervised or unsupervised SOM obtained from functions som, xyf or bdk.
- newdata: Data matrix, with rows corresponding to objects.
- whatmap: For supersom maps: the layers to take into account.
- weights: For supersom maps: weights of the layers that are used for mapping.
- scale.distances: whether to rescale distances per layer in the case of supersom maps (default): if TRUE the maximal distance of each layer equals one. If the absolute values of the distances per layer should be used, this argument should be set to FALSE. Note that in that case, when mapping the training data, the result returned by map.kohonen will differ from the mapping present in the map.
- ...: Currently ignored.
return a list with elements:
- unit.classif: a vector of units that are closest to the objects in the data matrix.
- dists: distances (currently only Euclidean distances) of the objects to the units.
- whatmap,weights,scale.distances: Values used for these arguments.
Function unit.distances
[edit | edit source]Calculate distances between units in a SOM [7].
unit.distances(grid, toroidal)
the arguments are:
- grid: an object of class somgrid.
- toroidal: if true, edges of the map are joined so that the topology is that of a torus.
return:
- Returns a (symmetrical) matrix containing distances. When grid$n.hood equals "circular", Euclidean distances are used; for grid$n.hood is "square" maximum distances. If toroidal equals TRUE, maps are joined at the edges and distances are calculated for the shortest path.
Function tricolor
[edit | edit source]Function provides colour values for SOM units in such a way that the colour changes smoothly in every direction [7].
tricolor(grid, phis = c(0, 2 * pi/3, 4 * pi/3), offset = 0)
the arguments are:
- grid: An object of class somgrid, such as the grid element in a kohonen object.
- phis: A vector of three rotation angles. Values for red, green and blue are given by the y-coordinate of the units after rotation with these three angles, respectively. The default corresponds to (approximate) red colour of the middle unit in the top row, and pure green and blue colours in the bottom left and right units, respectively. In case of a triangular map, the top unit is pure red.
- offset: Defines the minimal value in the RGB colour definition (default is 0). By supplying a value in the range [0, .9], pastel-like colours are provided.
return:
- Returns a matrix with three columns corresponding to red, green and blue. This can be used in the rgb function to provide colours for the units.
View
[edit | edit source]After the training phase, one can use several plotting functions for the visualisation; the package can show where objects are mapped, has several options for visualizing the codebook vectors of the map units, and provides means to assess the training progress. Summary functions exist for all SOM types. Furthermore, one can easily project new data into the trained map; this provides possibilities for property estimation [3].
Functions summary and print
[edit | edit source]Summary and print methods for kohonen objects. The print method shows the dimensions and the topology of the map; if information on the training data is included, the summary method additionally prints information on the size of the data and the mean distance of an object to its closest codebookvector, which is an indication of the quality of the mapping [7].
## S3 method for class 'kohonen': summary(object, ...) ## S3 method for class 'kohonen': print(x, ...)
the arguments are:
- x, object: a kohonen object
- ...: Not used.
return
Figure 2: Information retorned by function print about wine data
Figure 3: Information retorned by function summary about wine data
Function plot.kohonen
[edit | edit source]Plot self-organising map, obtained from function kohonen. Several types of plots are supported [7].
## S3 method for class 'kohonen':
plot(x, type = c("codes", "changes", "counts", "dist.neighbours", "mapping", "property", "quality"), classif = NULL,
labels = NULL, pchs = NULL, main = NULL, palette.name = heat.colors, ncolors, bgcol = NULL, zlim = NULL, heatkey = TRUE,
property, contin, whatmap = NULL, codeRendering = NULL, keepMargins = FALSE, heatkeywidth = .2, ...)
the arguments are:
- x: kohonen object.
- type: type of plot.
- classif: classification object, as returned by predict.kohonen, or vector of unit numbers. Only needed if type equals "mapping" and "counts".
- labels: labels to plot when type equals "mapping".
- pchs: symbols to plot when type equals "mapping".
- main: title of the plot.
- palette.name: colors to use as unit background for "codes", "counts", "prediction", "property", and "quality" plotting types.
- ncolors: number of colors to use for the unit backgrounds. Default is 20 for continuous data, and the number of distinct values (if less than 20) for categorical data.
- bgcol: optional argument to colour the unit backgrounds for the "mapping" and "codes" plotting type. Defaults to "gray" and "transparent" in both types, respectively.
- zlim: optional range for color coding of unit backgrounds.
- heatkey: whether or not to generate a heatkey at the left side of the plot in the "property" and "counts" plotting types.
- property: values to use with the "property" plotting type.
- contin: whether or not the data should be seen as discrete (i.e. classes) or continuous in nature. Only relevant for the colour keys of plots of supervised networks.
- whatmap: For supersom maps and a "codes" plot: what maps to show.
- codeRendering: How to show the codes. Possible choices: "segments", "stars" and "lines".
- keepMargins: if FALSE (the default), restore the original graphical parameters after plotting the kohonen map. If TRUE, one retains the map coordinate system so that one can add symbols to the plot, or map unit numbers using the identify function.
- Heatkeywidth: width of the colour key; the default of 0.2 should work in most cases but in some cases, e.g. when plotting multiple figures, it may need to be adjusted.
- ...: other graphical parameters, e.g. colours of labels, or plotting symbols, in the "mapping" plotting type.
Several different types of plots are supported:
- "changes": shows the mean distance to the closest codebook vector during training.
- "codes": shows the codebook vectors.
- "counts": shows the number of objects mapped to the individual units. Empty units are depicted in gray.
- "dist.neighbours": shows the sum of the distances to all immediate neighbours. This kind of visualization is also known as a U-matrix plot. Units near a class boundary can be expected to have higher average distances to their neighbours. Only available for the "som" and "supersom" maps, for the moment.
- "mapping": shows where objects are mapped. It needs the "classif" argument, and a "labels" or "pchs" argument.
- "property": properties of each unit can be calculated and shown in colour code. It can be used to visualise the similarity of one particular object to all units in the map, to show the mean similarity of all units and the objects mapped to them, etcetera. The parameter property contains the numerical values.
- "quality": shows the mean distance of objects mapped to a unit to the codebook vector of that unit. The smaller the distances, the better the objects are represented by the codebook vectors.
return:
Figure 4: Left Plot the function was called with type "counts". Right Plot the function was called with type "quality" [3].
Figure 5: The function Plot was called with type "property" [3].
Figure 6: The function Plot was called with type "codes" [3].
Figure 7: The function Plot was called with type "mapping" [3].
Figure 8: The function Plot was called with type "changes" [3].
Case Study
[edit | edit source]In this section, we illustrate a case study using package Kohonen.
Scenario
[edit | edit source]The standard form of self-organizing maps is implemented in function som. To map the 177-sample wine data set to a map of five-by-four hexagonally oriented units, the som function can be used. First, we load the package (from now on, we assume the package is loaded), and then the data, which are subsequently autoscaled because of the widely different ranges (especially the proline concentration, variable 13, deviates). The fourteenth variable is a class variable and is not used in the mapping; it will be used later for visualisation purposes [3].
Input data
[edit | edit source]As the input data we use de dataset wine that are included in the kohonen package. The dataset containing 177 rows and thirteen columns; object vintages contains the class labels. For compatibility with older versions of the package, variable wine.classes is retained, too. These data are the results of chemical analyses of wines grown in the same region in Italy (Piedmont) but derived from three different cultivars: Nebbiolo, Barberas and Grignolino grapes. The wine from the Nebbiolo grape is called Barolo. The data contain the quantities of several constituents found in each of the three types of wines, as well as some spectroscopic variables [3].
Execution
[edit | edit source]The following code can be use to create the map from the dataset. Note that first of all, you have to load the package and then load the dataset.
> library("kohonen")
Loading required package: class
> data("wines")
> wines.sc <- scale(wines)
> set.seed(7)
> wine.som <- som(data = wines.sc, grid = somgrid(5, 4, "hexagonal"))
> plot(wine.som, main = "Wine data")
Output
[edit | edit source]The result is shown in Figure 9. The codebook vectors are visualized in a segments plot, which is the default plotting type. High alcohol levels, for example, are associated with wine samples projected in the bottom right corner of the map, while color intensity is largest in the bottom left corner [3].
Figure 9: A plot of the codebook vectors of the 5-by-4 mapping of the wine data [3].
Analysis
[edit | edit source]The result of the training, the wine.som object, is a list. The most important element is the codes element, which contains the codebook vectors as rows. Another element worth inspecting is changes, a vector indicating the size of the adaptions to the codebook vectors during training. This can be used to assess whether the number of iterations is sufficient [3].
Extra
[edit | edit source]An example using the NIR data included in the package is shown below: for every ternary mixture, we have a nearinfrared spectrum, as well as concentrations of the three chemical compounds (summing to 1). Moreover, every sample is measured at five different temperatures. The aim in the example below is to model the water content (the second of the three concentrations). Of the three chemicals, water has the largest effect on the NIR spectra. We start by loading the data and attaching the data frame so that objects spectra, composition and temperature become directly available. Parameter xweight indicates how much importance is given to X; here it is set to 0.5 (X and Y are equally important), also the default value in xyf [3].
> data("nir")
> attach(nir)
> set.seed(13)
> nir.xyf <- xyf(data = spectra, Y = composition[,2], xweight = 0.5, grid = somgrid(6, 6, "hexagonal"))
> par(mfrow = c(1, 2))
> plot(nir.xyf, type = "counts", main = "NIR data: counts")
> plot(nir.xyf, type = "quality", main = "NIR data: mapping quality")
This leads to the output shown in Figure 4. In the left plot, the background color of a unit corresponds to the number of samples mapped to that particular unit; they are reasonably spread out over the map. Four of the units are empty: no samples have been mapped to them. The right plot shows the mean distance of objects, mapped to a particular unit, to the codebook vector of that unit. A good mapping should show small distances everywhere in the map [3].
References
[edit | edit source]- ↑ a b c d e f Pang-Ning Tan, Michael Steinbach and Vipin Kumar. Introduction to Data Mining. Addison Wesley; US ed edition. May 12, 2005.
- ↑ Katharine Mullen and Ron Wehrens.CRAN Task View: Chemometrics and Computational Physics.2010-11-02. URL [1].
- ↑ a b c d e f g h i j k l m n o p Ron Wehrens and Lutgarde M. C. Buydens. Self- and super-organizing maps in r: The kohonen package. Journal of Statistical Software, 21(5):1-19, 10 2007. URL [2].
- ↑ Frank, A. & Asuncion, A. (2010). UCI Machine Learning Repository [3]. Irvine, CA: University of California, School of Information and Computer Science.
- ↑ Wülfert F, Kok WT, Smilde AK (1998). Influence of Temperature on Vibration Spectra and Consequences for Multivariate Models. Analytical Chemistry, 70, 1761–1767.
- ↑ Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998). Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces Cerevisiae by Microarray Hybridization. Molecular Biology of the Cell, 9, 3273–3297.
- ↑ a b c d e f g h i j k l Ron Wehrens and Lutgarde M. C. Buydens. Self- and super-organizing maps in r: The kohonen package. Journal of Statistical Software, 21(5):1-19, 10 2007.