Structural Biochemistry/VDAC-1 and Analysis Using Solution NMR
Overview[edit | edit source]
Mitochondria have been known to be the powerhouse of the eukaryotic cell, possessing the ability to produce ATP which is used as cellular energy for the cell. However, mitochondria also fulfill other roles within the cell such as in metabolic pathways, apoptosis, cellular differentiation, and control of the cell cycle. As a result, to these multiple functions, mitochondria have evolved to develop a double membrane that surrounds the mitochondria complex. This double membrane functions as a high-traffic zone for the cell, possessing the ability to control what molecules go into the mitochondrion and what have to go out. For example, low-energy metabolites such as ADP have to go inside while high-energy metabolites such as ATP have to go out. This function of funneling ADP into mitochondria and ATP out of mitochondria is controlled by an integral membrane protein known as the voltage-dependent ion channel (VDAC), or also referred to as the mitochondrial porin.
The structure of VDAC has been examined for quite some time after it was discovered in 1975. Many structures of VDAC were determined, but the spatial arrangement, the topology, of the structure for the beta-strand could not be determined. However, in 2008, three long-term efforts to determine the three-dimensional structure of VDAC-1 were determined at atomic resolution. Three structures of the isoform VDAC-1 were determined by different methods. One was determined by using NMR spectroscopy alone, another by X-ray crystallography alone, and the last one using a combination of both NMR spectroscopy and X-ray crystallography. The comparison of these three different structures of VDAC-1 is examined as well as the discussion of the importance of solution NMR to determine the structure of VDAC-1.
Structure of VDAC-1 and Comparison of Three Structures[edit | edit source]
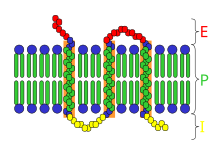
The structure of VDAC-1 is very unique as it contains a very large beta-barrel. For all three structures, the number of strands in this beta-barrel and the spatial arrangements of molecules is the same. In studying the amino acid sequence of VDAC, it has been identified as being conserved from yeast to human. As a result, the overall folding pattern of the structure is known to be the same in all eukaryotes. In the three structures of VDAC-1, one of the structures is derived from a mouse while the other two structures are derived from humans. When comparing the mouse form of VDAC-1 to the human form of VDAC-1, the two forms are highly identical, differing in only four amino acids. Due to the very small changes in amino acid sequence between the mouse and human forms, the three-dimensional folded structures are very similar. To further confirm the beta-barrel structure of VDAC-1, denaturation of the VDAC-1 protein was performed to allow it to refold into the detergent LDAO. The refolded VDAC-1 structure was then placed into a different environment containing bicelles known as DMPC. By placing the refolded VDAC structure in a different solution environment, the same beta-barrel structure was observed again, and it was concluded that this beta-barrel structure of VDAC-1 is the same no matter what type of environment solution it is placed in.
The beta-barrel structure of VDAC-1 is fairly unique because it is the only structure that is observed in any eukaryotic membrane protein, and it is also the only known beta-barrel membrane protein that contains an odd number of strands. The rest of the beta-barrel proteins are observed to arrange into anti-parallel beta sheets, and because of this, an even amount of strands is needed to stabilize the entire beta-sheet structure through hydrogen bonding. It is unknown why a beta-barrel structure is stabilized with an odd number of strands as the folding mechanism of this protein is not fully understood. The structure of the beta-barrel is defined using two numbers that are the number of strands, n, and the shear number, S. The shear number in the beta-barrel can be identified as the pairs of alpha-carbon atoms in adjacent strands that lie on a helical trace across the surface of the beta-barrel. The side chains of the alpha-carbon atoms must be pointed to the same side of the sheet, and following the trace of the helix once around until it arrives back at the first strand a certain number of residues away from the starting point is known as the shear number of the beta-barrel. In beta-barrels, the shear number is always even in order to have the hydrophobic residues of the protein on the outside of the complex. Beta-barrel structures usually contain a shear number in the range of n and n+4.
Another comparison that is made to differentiate the three VDAC-1 structures is the residues that branch off the protein that is not part of the beta-barrel. The 1-23 residues are compared between each of the three structures, but through the use of NMR, only residues 6-10 have been identified to be an alpha-helical structure. Also, through the use of X-ray crystallography, the structure of mouse VDAC-1 was observed to contain three aliphatic residues: Leucine 10, Valine 143, and Leucine 150. From the crystallized structure, it was observed that Valine 143 and Leucine 150 are the only hydrophobic side chains that point to the barrel interior from the barrel wall. Residues 11-20 in the mouse structure and human structure appear to contain similar segments. However, the conformation of these segments differs between these two structures. Both structures were analyzed at cryogenic temperatures through the use of NMR. The conformational changes between these structures are exposed through the use of NMR because as the conformations change, residues of the proteins will end up interacting with other different neighboring residues. As a result, these conformational changes can lead to multiple resonance lines, reduced signal intensity, or line broadening on the NMR graphs for the structures.
Solution NMR in Determining Structure of VDAC-1 and Other Integral Membrane Proteins[edit | edit source]
In determining the beta-barrel structure of VDAC-1, researchers have stated that the combination of NMR and X-ray crystallography data were not enough in fully determining the structure. As a result, the use of solution NMR techniques was used instead to solve the beta-barrel structure of this membrane protein. In total, nine structures of integral membrane proteins have been solved using solution NMR. In using solution NMR, two important techniques are used in determining membrane protein structures such as VDAC-1: protein refolding and deuteration of the detergent micelle.
The use of protein refolding from a denatured state has a very low success rate for most membrane proteins, but if the refolding process is successful, there are many benefits that help to study the structure of membrane proteins with much greater ease. First, the process of protein refolding can lead to a very high yield of the newly folded structure of the protein. In the case of VDAC-1, an average of 40 mg of VDAC-1 was obtained in a 1 liter solution of an E. coli cell culture. Second, protein refolding helps to purify the membrane protein to an extremely high degree. This is extremely important in studying the structure of VDAC-1 as the data obtained from X-ray crystallography and NMR would be accurate in examining the true structure. Third, protein refolding has a high reproducibility, which goes together with the high purity. Fourth, because high yield and high reproducibility can be done from protein refolding, efficient perdeuteration and selective isotope labeling can be done. Finally, since the predeuterated protein goes through a denatured state in the in deuterated-water, all the amide compounds are readily protonated by the deuterated-water, and therefore, the structure of proteins such as VDAC-1 are much easier to identify due to the presence of D’s instead of H’s.
The use of deuterated detergent in solution NMR is the second technique that helps in identifying the structure of large membrane proteins such as beta-barrel of VDAC-1. From other NMR studies, compounds not placed in deuterated solutions produce very broad resonance lines due to the strong dipole-dipole interactions between different atoms, causing the spectral sensitivity to be reduced an extremely significant amount. By examining membrane proteins such as VDAC in deuterated solutions, a much more specific NMR graph is observed. For example, when using the Nitrogen-15-resolved-NOESY spectra, when a deuterated detergent was replaced with a protonated detergent, a decrease of 10-30% in sensitivity was observed. This decrease in sensitivity is clearly seen when analyzing the spectra of the methyl groups of the aliphatic residues of Isoleucine, Leucine, and Valine. The NOESY spectra of these groups did not produce a clear spectra identifying these compounds in a protonated detergent, but when a deuterated detergent was used, clear images of these groups were able to be identified, concluding that the use of solution NMR in a deuterated detergent proved to be a powerful method in determining the structures of integral membrane proteins such as VDAC-1.
References[edit | edit source]
Hiller, S. The role of solution NMR in the structure determinations of VDAC-1 and other membrane proteins. 2009, Current Opinion in Structural Biology. p. 396-401.