Structural Biochemistry/Nucleic Acid/RNA/RNA Interference

Discovery[edit | edit source]
RNAi was first introduced when plant biologists attempted to introduce genes into a petunia. When they added a gene that attempted to deepen the flowers purple color, the gene actually inhibited it. The resulting flowers had white patches or were completely white.
Soon after this discovery, another group of researchers realized that this same gene-silencing phenomenon was occurring in experiments with C. elegans. These scientists figured out that RNAi is triggered by double-stranded RNA, which is not typically found in healthy cells. Two well known scientists, Andrew Fire and Craig Mello, were awarded the 2006 Nobel Prize in physiology or medicine for this discovery. [1]
Biological Implications[edit | edit source]
RNA interference (RNAi) is a natural mechanism within the cell used to silence the expression of certain genes. Small RNA molecules play essential roles in regulating gene expression by RNA interference. There are three basic characteristics of these pathways:
1) Small RNA biogenesis
2) Formation of RNA-induced silencing complexes.(RISCs)
3) Targeting of complementary mRNAs.
RNA interference is triggered by the enzyme Dicer, which cleaves long double-stranded RNA (dsRNA) producing 20 to 30 nucleotide RNAs whose sequences can base pair with segments of mRNA transcripts. Then the newly generated microRNAs (miRNAs) or small interfering RNAs (siRNAs) will assemble into complexes designed to complementarily fit into target RNA strands that wish to be silenced. The induced silencing complexes are called RNA- induced silencing complexes (RISC) and are constructed into large multiprotein effectors, called RNA-induced silencing complexes (RISCs), which bind to target transcripts and trigger their destruction.
Cognate RNA is then cleaved in the middle region bound to the siRNA strand. This mechanism has been theorized to have a self-defense purpose to protect cells against viral infections or cancerous cells.
RNAi can help to study tissue regeneration. RNAi shuts down individual genes during the tissue regeneration and the scientists can understand what genes in amphibians are involved in regenerating tissue when missing limbs are regrown. By understanding this process, they hope to learn how to regenerate human tissue.
Origin[edit | edit source]
RNAi is proposed to have evolved about a billion years ago, before plants and animals diverged. This is due to the fact that it exists in all living organisms, from plants to animals.
Modern hypotheses state that RNAi evolved as a cellular defense mechanism against invaders such as RNA viruses. When they replicate, RNA viruses temporarily produce a double-stranded form. This double-stranded intermediate would trigger RNAi and inactivate the virus’ genes, preventing an infection.
RNAi may also have evolved to combat the spread of genetic elements called transposons within a cell’s DNA. Transposons can wreak havoc by jumping from spot to spot on a genome, sometimes causing mutations that can lead to cancer or other diseases. Like RNA viruses, transposons can take on a double-stranded RNA form that would trigger RNAi to clamp down on the potentially harmful jumping. [2]
Cellular Mechanism[edit | edit source]

RNAi is a process in which RNA is used to scilence genes. the main player in this process is the RNA-induced silencing complex (RISC), the complex is activated by short double-stranded RNA molecules. [3]. dsRNA can come from infection by a retro virus or artificially inserted (exogenous), the RNA can also come from within the cell's own genome (endogenous). [4].
dsRNA[edit | edit source]
There are two approaches toward dsRNA depending on its origin, whether it be exogenous (from outside the cell) or endogenous (from inside the cell).
-Exogenous: the foreign dsRNA is detected and bound by an effector protein, the protein initiates the dicer to cut up the dsRNA, the same effector protein helps in transporting the siRNA to RISC.[5]
-Endogenous: the target dsRNA is cut up by the Dicer into single stranded siRNA, which are then transported to an active RISC. When they are incorporated into the RISC the siRNA base pair with their corresponding sequences on mRNA strands, which are then cleaved at those sites. By cleaving the mRNA the synthesis of protein is halted [6].

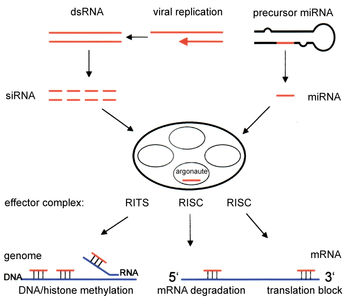
RISC[edit | edit source]
RISC stands from RNA-induced silencing complex, its active components consist of endonucleases and argonaute proteins. The function of each respectively is to recognize a complimentary sequence on mRNA (complementary to the sequence of the bound siRNA) strands and cleave the mRNA (argonaute proteins). This process is ATP independent and act directly through components of the RISC [7] [8].
How RNA pairs with the argonaute protein, structurally was determined by X-ray crystallography, through x-ray crystallography the active sites were determined which led to accurate information regarding how the RNA binds to the argonaute protein. In the active site, the phosphorylated 5' end of the RNA strand enters and bonds with a cation (i.e. magnesium) and by having an aromatic stacking structure between the 5' nucleotides in the siRNA. It has been inferred that the active site contains the ability to pair the siRNA with its corresponding mRNA. [9].
Research Implications[edit | edit source]
RNAi-based therapies have been proposed as a way to regulate and get rid of several disease causing genes. This path has shown to be the most promising. A good target for this type of therapy would be all forms of cancer. Cancer is often caused by overactive genes and regulating the activity of these could stop the spread of it.
Viral infections are also hypothetical targets for RNAi therapies. Many believe that RNAi actually evolved as a way to combat RNA viruses. Reducing the expression of important viral genes would leave the virus helpless and prone to attack by the immune system. In vitro, studies have already indicated that HIV, polio, HCV and others have been reduced by these therapies.
RNAi is already serving as a way to identify function of certain genes. Prior to this discovery, researchers had often resorted to inserting new genes into an organism to see what the effect would be. More recently however, scientists can merely silence the gene of interest and observe the effects that the target gene has on organism function. It can also shed light upon complex cellular pathways.
RNAi has been a novel and highly important discovery for research. For years, scientists had been intensely studying how proteins regulate gene activity, focusing most of their attention on proteins called transcription factors. Now RNA, through RNAi and related processes, is known as an essential player in the cell’s complex technique of gene regulation. [10]
Research Applications[edit | edit source]
As mentioned in the section above, RNAi can be used to selectively “silence” targeted genes in order to analyze the affects this will incur on the model organism. One area of research focuses on regeneration, the regrowth of lost or damaged body parts. This ability is quite common in nature. For example, tree stumps can grow sprouts that develop into new stems, leaves, and flowers; in lab, a mass of undifferentiated cells can grow into a mature plant; in fact, a section of certain plants composed of fully-differentiated cells can also grow into a mature plant. Animals, too, have regenerative abilities: including invertebrates such as sponges, hydra, planarians, and starfish, as well as vertebrates such as salamanders and amphibians. Humans, on the other hand, have only limited regenerative abilities. Apart from healing wounds, humans can regenerate some of the liver and the tips of fingers and toes [11]. Wouldn’t it be amazing if scientists could find a key to regenerating human tissues? RNAi is currently being used to target specific genes and turn them off in planarians and amphibians in order to analyze the functions of those genes. This way, researchers hope to find out which genes are responsible for regeneration.
Role in Tissue Regeneration[edit | edit source]
RNA interference (RNAi) is a mechanism that organisms use to silence genes when their protein products are no longer needed. The silencing happens when short RNA molecules bind to stretches of mRNA, preventing translation of the mRNA. To focus in on the genes that enable planarians to regenerate, Sánchez Alvarado and his coworkers are using RNA interference (RNAi). RNAi is a natural process that organisms use to silence certain genes. Sánchez Alvarado’s group harnesses RNAi to intentionally interfere with the function of selected genes. The researchers hope that by shutting down genes in a systematic way, they’ll be able to identify which genes are responsible for regeneration. The researchers are hoping that their work in planarians will provide genetic clues to help explain how amphibians regenerate limbs after an injury. Finding the crucial genes and understanding how they allow regeneration in planarians and amphibians could take us closer to potentially promoting regeneration in humans.
Specifically in planarians, which can regrow a whole worm from a small fraction of its body, RNAi’s ability to shut off specific genes has led to the discovery that the location of head and tail formation is controlled by *hedgehog signaling and the Wnt/B-catenin pathway. The Wnt/B-catenin pathway regulates the formation of the anterior-posterior axis. “Silencing” either hedgehog or Wnt/B-catenin with RNAi causes head and tail to grow at wrong ends [12]. In addition, some basic researchers are trying to figure out how stem cells work by planarians. These worms are like stem cells in the sense that they can regenerate. Planarians’ resemblance to stem cells isn’t just coincidence. Scientists have discovered that planarians can perform the amazing act of regeneration due to the presence of specialized stem cells in their bodies. Developmental biologist Alejandro Sánchez Alvarado of the University of Utah School of Medicine in Salt Lake City used the gene-silencing technique RNAi to search for planarian genes that were essential for regeneration. He found 240 genes that caused a physical defect in the worm’s growth and regenerative ability when silenced. Interestingly, 16 percent of these looked very much like genes that had been linked to human disease.
- in addition to regeneration in planarians, hedgehog signaling is vital to brain, intestinal tract, finger, and toe development in mammals.
RNAi and Neurological Diseases[edit | edit source]
RNAi not only protects cells from foreign genes, it is also involved in regulating the cells own genes, including the cell’s own set of noncoding mRNA’s. Thus, improperly functioning RNAi can lead to diseases and inherited disorders, including fragile X syndrome. Fragile X causes mental retardation because of the loss of FMRP, a protein usually synthesized from the FMR1(fragile X mental retardation 1) gene. It was discovered that FMRP is a component of RISC, indicating that the loss of this protein prevents RNAi in neurons from functioning properly, thus causing mental retardation [13]. (More research needs to be done to establish this link.)
On the flip side, RNAi has the potential to treat neurological diseases as well. In a similar fashion to how RNAi eliminates foreign mRNA from viral infections, the high specificity of RNAi can be used to target mutations of normal genes that lead to neurological diseases. This way, RNAi can mediate the effects of detrimental dominant alleles by “knocking out” expression of these mutant genes while leaving normal ones alone. Accordingly, this potential can be expanded to other diseases including those caused by triplet expansion or trinucleotide repeats (Neurodegernative diseases such as Spinobulbar Muscular Atrophy and Hungtington’s in addition to Fragile X). [14]
References[edit | edit source]
- ↑ Macrae I, Zhou K, Li F, Repic A, Brooks A, Cande W, Adams P, Doudna J (2006). "Structural basis for double-stranded RNA processing by dicer". Science. 311 (5758): 195–8. doi:10.1126/science.1121638. PMID 16410517.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Hammond S, Bernstein E, Beach D, Hannon G (2000). "An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells". Nature. 404 (6775): 293–6. doi:10.1038/35005107. PMID 10749213.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ U.S. Department of Health and Human Services. Inside the Cell. September 2005.<http://www.nigms.nih.gov>.
National Institute of General Medical Sciences [15]
Biology Pages [16]
Functional Genomics, Fragile X Syndrome, and RNA Interference [17]
The New Genetics (2006): n. pag. U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES, National Institutes of Health, National Institute of General Medical Sciences. Web. <http://www.nigms.nih.gov>.