Structural Biochemistry/Antivirals
Introduction of antivirals[edit | edit source]
Antiviral drugs were invented to prevent viral infections. They are one class of antimicrobials and harmless to human body. Instead of destroying the target pathogen, antiviral drugs inhibit proteins that contribute one or several steps in viral infection. It is difficult to invent new safe and effective antivirals because the harm to host cells must be taken into consideration and variation of viruses.
Four classes of antivirals:
- Neuraminidase inhibitors
- Protease inhibitors
- Neutralizing antibodies
- Protein-based fusion inhibitors
Neuraminidase Inhibitors[edit | edit source]
Introduction of Neuraminidase[edit | edit source]
Neuraminidase, also called sialidases, is glycoside hydrolase enzymes, which can cleave the glycosidic linkages of neuraminic acids.
There are nine subtypes of neuraminidase. Three major classes of neuraminidase enzymes are:
- Viral neuraminidase
- Bacterial neuraminidase
- Mammalian neuraminidases:
sialidase 1 (lysosomal sialidase), with symbol NEU1.
sialidase 2 (cytosolic sialidase), with symbol NEU2.
sialidase 3 (membrane sialidase), with symbol NEU3.
sialidase 4, with symbol NEU4.
In order to better understand the process of developing neuraminidase inhibitors, the case of influenza virus neuraminidase inhibitor development will be examined. The neuraminidase of influenza virus is a homotetrameric glycoprotein anchored to the viral membrane. It spreads on the membrane of influenza virus, helping mature influenza virus leave from host cell and infect new cells. For each influenza virus, there are near 100 neuraminidases spreading on its surface.

Introduction of neuraminidase inhibitors[edit | edit source]
Inhibiting neuraminidase keeps newly produced viruses in the host cell and prevents the infection of other cells.
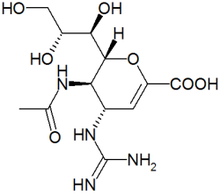

There are 2 kinds of influenza virus, type A and type B. Old drugs, such as amantadine, only kill type A influenza virus effectively; it is useless to type B influenza virus. The new antivirals are invented by changing the functional group on drugs to reduce the drug resistance and change their reactivity with type B influenza virus. They can inhibit both type A and type B. There are two drugs (Zanamivir, Oseltamivir) that are commercially available in the market now. The third is still in clinical evaluation.
Zanamivir is the first neuraminidase inhibitor commercially developed to treat influenza virus A and virus B. It was discovered in 1989 by scientists led by Mark von Itzstein. Zanamivir has molecules formula C12H20N4O7, and molecules mass 332.31 g/mol. There are no known toxic effects caused by Zanamivir.
Oseltamivir is an antiviral that used to treat and prevent influenza A virus and influenza B virus infection.It was approved in 1998 Feb. Oseltamivir has molecules formula C16H28N2O4, molecules mass 312.4 g/mol. It attacks the hydrophobic pocket in the active site of neuraminidase protein of influenza virus to stop the reproduction process of virus.
Protease Inhibitors[edit | edit source]
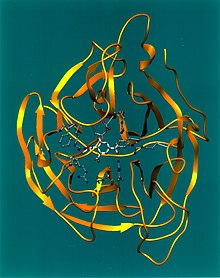
Protease is an enzyme that exists to cleave polyproteins. Specifically for the case of HIV protease, the enzyme is necessary to cleave the Gag and Pol polyproteins, which is vital to the maturation of HIV virus. The cleaved proteins are used to build components for infections HIV virions. By blocking this protease, inhibitors stop the reproduction process of HIV virus, causing HIV virions remain uninfectious. Designing a drug to inhibit this enzyme is difficult as there are dynamic properties of the protein that control its ability to bind to substrates. However, since HIV protease is very important to the life of the HIV virus, it is a viable option for designing new drugs.
Let’s take a look at the structure of HIV-1 protease first, which was found thanks to x-ray crystallography.

The HIV protease is a symmetric dimmer with a pair of active-site residues-catalytic aspartates. Moreover, these active sites are quite close to the symmetrical axis.
There are several drugs that are used to control HIV virus and treat AIDS:
Amprenavir was approved by FDA on April 15, 1999. Patients only need to take medicine twice a day, instead of three times. Amprenavir was discontinued to produce on 2004.

Tipranavir is a nonpeptidic protease inhibitor manufactured by Boehringer-Ingelheim, approved by FDA on June 22, 2005.

Darunavir is second-generation protease inhibitor, used to treat HIV infection. Darunavir was approved by FDA on June 23, 2006. It has molecules formula C27H37N3O7S, and molecules mass 547.665 g/mol.
Unfortunately, just like many other drugs, human immunodeficiency virus (HIV) protease inhibitors have also faced obstacles to due to resistance and bioavailability. Hence, it is rather appropriate to discuss about using phosphonic acid esters as prodrugs to approach this two important issues. The Gilead group, who have been working on improving the bioavailability of Zanamivir, have discovered a specific covalent structure of a phophonate diethyl ester to TMC-126, which is an analog of the protease inhibitors amprenair. One essential feature of the phosphonate diester is its cell permeability. After it passes through the cell membrane, the phosphonate diester is prone to go through intracellular enzymatic hydrolysis to yield diphosphonate dianion, which is less likely to diffuse out of the cell. As a result, this compound increases the effective concentration of the HIV protease inhibitors at the intracellular target site. On the other hand, the low resistance of the inhibitors can be ascribed to a change in the binding thermodynamics of the GS-8374 vis-`a-vis the nonphosphonate parent TMC-126. Solvent reorganization is a fairly important procedure in the process, as suggested by the energetic of the process, combined with the lack of significant interactions of the phosphonate with protease. The structure of phosphonate also gives the molecule positional flexibility in the active site, which decreases the probability of mutations. This concept may be applied to other HIV protease inhibitors as well, while solvent reorganization remains as an undetermined idea for other proteases and therapeutic areas. Inhibitors of HIV viral membrane, such as oligopeptide and enfuvirtide, are proved to be effective in acting as disruptors of helical bundle formation during the fusion process. However, the cost and dosing regimen of enfuvirtide have made it rather difficult to carry out therapy for multidrug-resistant HIV patients. Drug designers are now looking into solutions in addressing HIV patients’ resistance to enfuvirtide. Biologics of considerably-sized molecule that can compete effectively in binding are required to disrupt relatively large-surface protein-protein interactions, and this places limitations on the other aspects of the drug.[1]

Neutralizing Antibodies[edit | edit source]
Neutralizing antibodies are highly specific and highly effective as antiviruses. They can be introduced to the body through vaccinations, or due to earlier infections. Due to the specificity of antibodies, research has shown that in single amino acid changes in the antigen resulted in preventing the binding of antibodies to antigens. Structural studies following this research showed that the single amino acid changes resulted in only local structural changes.
Although antibodies are highly effective in targeting viruses specifically, research shows that viruses also have mechanisms in order to avoid detection by antibodies. For example, with influenza antigens, the enzymatic site is presented as smaller than the typical site targeted by antibodies. HIV gp160 uses carbohydrates and conformational changes in an attempt to mask the enzymatic site.
There is currently one monoclonal antibody that is registered as an antiviral agent: Palivizumab, or Synagis, is raised in mice and used in pediatrics for targeting the respiratory syncitial virus (RSV). In developing this antibody, three specific variants of RSV Fusion were selected for in vitro testing: K272M, K272Q, and N268I.
There also exists interest in using this type of therapy for hepatitis C virus, but further developments still must be made.
Protein-based Fusion Inhibitors[edit | edit source]
This type of inhibitor is based on preventing the fusion of the viral membrane to the cell membrane, thus preventing the virus from interacting with the cell altogether. By inhibiting this mechanism of viruses, cells would not be infected; therefore, this is a very desirable reaction to target for drug developers. The molecular bases for this reaction is developed from the influenza virus hemaglutanin, the fusion protein, in pre- and post-fusion conformations. Observations have found that the membrane fusion process follows the production of a six-helix bundle. The specific intermediate targeted in this pathway is a trimeric coiled coil in helical region 1 (HR1) where a "fusion peptide" found at the N-terminus associates with the target membrane.
The first drug developed through this approach is Enfuviritide, an oligopeptide with an sequence that overlaps the helical region 2 (HR2) sequence of the HIV fusion protein gp41. The goal of the drug is to prevent the formation of the helical bundle through competition, replacing the natural substrate and thus creating a barrier. Some studies show, however, that it only weakly inhibits the formation of the six helix bundle. It is possible that the mechanism of the drug is more complex than expected, and more studies must be conducted.
Design of New Antiviral Medicines[edit | edit source]
According to the article “New Antivirals and Drug Resistance” by Peter M. Colman, he announced a hypothesis that the increase of similarity between the drug and the target’s natural ligands (substrates) can make the virus more difficult to resist drugs. Besides of this factor, people should also take entropy compensation and solvent anchoring into consideration to design new and better antiviral medicines.
There are several ways to improve the drug by combating with drug-resistant virus:
- Raise the concentration of the drug: It would make the drug more effective if the drug can be distributed widely and also close to the target in an infected cell. Then the higher concentrated and more specifically distributed drug can cause the mutations less significant.
- Add a charged phosphonate to protease inhibitors: Occasionally, scientists find out that a phosphonate-containing inhibitor can bind to a wider range of viral-resistant variants. The example is GS-8373, which was analyzed by crystallography that the phosphonate groups remain their activities after mutation happens. Other parts without phosphonates loose 10- to 40-fold activity. However, according to the article “New Antivirals and Drug Resistance” by Peter M. Colman, scientists failed in raising virus in an environment with phosphonate-containing protease inhibitors.
- Increase drug’s similarity to the natural substrates: The similarity in shape, charge, and other configurations can improve drug’s effectiveness to bind to virus for inhabitation.
Reference[edit | edit source]
- ↑ Annu. Rev. Biochem. 2011. 80:239-46 The Annual Review of Biochemistry is online at biochem.annualreviews.org
1. Peter M. Colman, "New Antivirals and drug Resistance"
2.SHAO Huayi,LI Zhuorong, "Overviews of neuraminidase inhibitors"
3.http://baike.baidu.com/view/555631.htm
4.http://baike.baidu.com/view/344762.htm