Structural Biochemistry/Nucleic Acid/RNA/Ribosomal RNA (rRNA)
Ribosomal RNA, also known as rRNA, is a significant component of the ribosome. rRNA fabricates the polypeptides and provides a mechanism for decoding mRNA into amino acids and interacts with the tRNA during translation. rRNA was once known to be the key structural component of ribosomes, but its actually found to be a catalytic element for protein synthesis. It is the most abundant type of RNA (about 80%) in the cell.
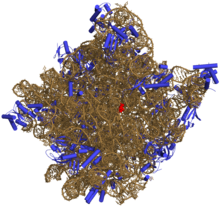
rRNA is comprised of a large and small subunit. Prokaryotic rRNA is 70 svedbergs large. A svedberg is a unit of measurement for the sedimentation coefficient or how fast the molecule sediments when centrifuged. The 70S rRNA contains a large 50S subunit which includes a 23S and 16S subunit and a small 30S subunit which contains a 5S subunit. The 23S, 16S and 5S units are essential during protein synthesis, and the structure and function of the ribosomes. The formation of these RNAs take place by cleaving the primary 30S subunits and processing further by folding the molecule to form internal base pair structures. Experiments involving chemical probing methods have been conducted which have provided a detailed model of the secondary structure of the 16S subunit. The secondary structure was obtained through analyzing and comparing the sequences. Proteins containing the 16S ribosomal rRNA can fold and form the 30S subunit.The conformational change of the 16S ribosomal rRNA plays a crucial role in the assembly of the ribosome. The 5S unit found in the 30S subunit is an important part of the large subunit of most ribosomes found in organisms. rRNA is the most abundant of the three major types of RNAs with a 80% relative amount in E. coli for example, following by tRNA (15%) and finally mRNA (5%). Ribosomal RNA has a mass of 1.2 x 10^3 kd and 3700 number of nucleotides in E. coli.
With the help of x-ray crystallographic technique, scientists are able to reveal the detailed features of secondary structures.
The use of Polymerase Chain Reaction (PCR) has been of great importance in the amplification of rRNA genes. PCR is used to amplify rRNA genes in many organisms, however, it is found that the amplification of rRNA genes via traditional PCR methods cannot be conducted in extremely thermophilic organisms.
rRNA contains two tRNA binding sites, an A site and a P site. At the A site, the rRNA binds to a aminoacyl-tRNA, a tRNA bound to an amino acid. The amino acid is transferred to a peptidyl-tRNA containing the growing peptide chain. After the amino acid is added, the empty tRNA is moved to the P binding site where it is ejected. The mRNA then shifts 3 bases (1 codon) for the next aminoacyl-tRNA to bind to the A binding site.
In prokaryotes, rRNA are formed by cleavage and other modifications of nascent RNA chains. Therefore precursors of transfer and ribosomal RNA are cleaved and chemically modified after transcription (DNA --> RNA) in prokaryotes.
Base Pairing[edit | edit source]
rRNA takes great part of base-pairing between the codon and the anticodon. "Adenine 1493, one of three universally conserved bases in 16S rRNA, forms hydrogen bonds with the bases in both the codon and the anticodon only if the codon and anticodon are correctly paired."
Referrence[edit | edit source]
M. Ogle and V. Ramakrishnan. Annu. Rev. Biochem. 74 (2005):129-177.