Proteomics/Protein Identification - Mass Spectrometry/Types Mass Spectrometry
This Section:
Types of Mass Spectrometry[edit | edit source]
Previous Section: Instrumentation
AMS (Accelerator Mass Spectrometry)[edit | edit source]
In AMS (Accelerator Mass Spectrometry), a “tandem accelerator” is used to accelerate the ions at several million volts. This tandem accelerator works twice. During the first stage, negative ions are created from an ion source. These negative ions are accelerated by an electrostatic accelerator to very high kinetic energies. Then a sheet of carbon is introduced in to the instrument, to convert the negative ions into positive ions. Since these ions are now positive, they accelerate away from the center rapidly. Now a velocity selector with a combination of both electric and magnetic fields is used to allow the ions with particular mass and kinetic energy through. Finally m/z is determined by passing through a magnetic or electric sector mass analyzer.
Gas Chromatography-MS[edit | edit source]
Gas chromatography separates the components of a mixture and mass spectroscopy characterizes each of the components individually. This method is a preferred method within the proteomics community, and is known as a "golden standard" for analysis.[[1]] In gas chromatography (GC), the mobile phase is an inert gas such as helium. Gas chromatography is a method in which molecules are separated based on volatility and bond characteristics when subjected to a carrier gas. The sample enters a vacuum chamber through an inlet and is converted into gas phase ions. The ions are sorted according to their mass to charge ratios, usually with a quadrupole or ion trap. This data is converted into representative electrical signals and finally, a mass spectrum is created.

This technique is a simple type of chromatography that is coupled to a mass spectrometer. When coupled with MS it is known as a hyphenated analytical technique due to the combination of two separately performed techniques. Dependent on the type of chromatography, buffers, and conditions, some methods of separations can be coupled with MS and some cannot. GC is a method that can normally be coupled with MS due to the samples already being in a volatile form, the form that MS can only read in. Due to the lack of buffers and salts this method does not interfere with the MS detection ability.
A great advantage to this method is that a researcher can take an organic solution, inject it into the instrument, separate the individual components, and identify each of them. Furthermore, the researcher can determine the quantities (concentrations) of each of the components.
There are some important limitations to GC-MS:
If the GC instrument does not separate the samples compounds completely or correctly, the MS feed is impure. If this occurs, background noise can appear in the mass spectrum resulting in poor resolution and poor retention times which can cause significant error in data.
Of course, the MS portion of the coupling involves a large amount of analysis from a skilled technician. Many aspects come into play, and the analyst must correlate computer calculations with system conditions. This can produce a large amount of human error within the data.
Liquid Chromatography-MS[edit | edit source]

LC-MS separates compounds chromatographically before they are introduced to the ion source and mass spectrometer by means of using liquid mobile phases which ultimately must be volatilized before entering the MS. It differs from GC-MS in that the mobile phase is liquid, usually a combination of water, organic solvents, and samples instead of gases. The method of coupling high performance liquid chromatography (HPLC) can also be performed with MS. A HPLC simply uses a smaller column that is highly chemically modified to separate on a more precise level than normal LC. Once a sample is injected it goes through a column which separates it based on charge and goes into a drying chamber where the sample is volatilized by a drying gas such as N2. The ions are then collected into a gas capillary where they are collected to be injected further in the system. When the ions proceed out of the gas capillary, the ions go through an area where collision activated dissociation occurs between a skimmer and the capillary, causing the ions to exit individually. The area where the gas capillary and the skimmer meet is the area where volatilization begins. From the capillary, the liquid ions are put through a "Taylor cone". The Taylor cone creates the effect of a fine filament of liquid that volatilizes into a gaseous form by changing its stable liquid droplets to unstable liquid droplets before changing them to gas phase ions. The samples then proceed to an inlet for the mass spectroscopy machine into a quadrupole where they are further separated by charge to mass and then moved to a detector to obtain a mass spectrum.
ICP-MS (Inductively Coupled Plasma-Mass spectrometry )[edit | edit source]
ICP-MS involves the formation of gas containing electrons, ions and neutral particles from Argon gas. The sample is atomized and ionized by this gas. In a high vacuum mass analyzer, these ionized atoms from gas are passed through cones (apertures). Based on the m/z ratio, the isotopes of the elements can be identified. The intensity of the element’s peak in the mass spectrum will be proportional to the amount of the isotope in the original sample[2].
IRMS (Isotope Ratio Mass Spectrometry)[edit | edit source]
Isotope ratio mass spectrometery (IRMS) is used to measure mixture of stable isotopes. It has two inlets that help in repetitive measurements with continuous supply of sample gases. This supply is sequentially switched over by a valve. This instrument has collector with conductive metal vessels that neutralizes ions, also multicollectors in some of them allowing detection of multiple isotopes. Samples that are introduced in to this instrument should be pure gases obtained from combustion or chemical trapping. An isotopic make up of a sample can be obtained by comparing the standard isotopes to the detected isotopes from the sample.
Ion Mobility Spectrometry-MS[edit | edit source]
Previously it was believed that this method could not be coupled with MS due to its non-volatile state as well as reaction of buffers, however a few groups have been experimenting with techniques and methods to couple these two performances to be able to analyze simple samples. Experiments and current tests are attempting to couple these two by using a curtain gas from the MS as a drift gas for the ion mobility to allow proper gaseous form for the MS to run. [3] This method is believed to be a fast and cheaper alternative to the slower expensive methods of separations for those samples that do not require much resolution.
The design of the ion mobility spectrometer allows reasonably fast installation (about 1 hour), and thus the ion mobility spectrometer can be considered as an accessory of the mass spectrometer. The ion mobility spectrometer module can also be used as an independently operated device when equipped with a Faraday cup detector. The drift tube of the ion mobility spectrometer module consists of inlet, desolvation, drift, and extraction regions. The desolvation, drift and extraction regions are separated by ion gates. The inlet region has the shape of a stainless steel cup equipped with a small orifice. Ion mobility spectrometer drift gas is introduced through a curtain gas line from an original flange of the mass spectrometer. After passing through the drift tube, the drift gas serves as a curtain gas for the ion-sampling orifice of the ion mobility spectrometer before entering the ion source. Counter-flow of the drift gas improves evaporation of the solvent from the electrosprayed sample. Drift gas is pumped away from the ion source through the original exhaust orifice of the ion source. Initial characterization of the ion mobility spectrometer device includes determination of resolving power values for a selected set of test compounds, separation of a simple mixture, and comparison of the sensitivity of the electrospray ionization ion mobility spectrometry/mass spectrometry (ESI-IMS/MS) mode with that of the ESI-MS mode. (Sysoev, A 2004)
MALDI-TOF[edit | edit source]
Matrix Assisted Laser Desorption Ionisation (MALDI) (F. Hillenkamp, 1991) deals with thermolabile, non-volatile organic compounds and those of high molecular mass. It is used in for the analysis of proteins, peptides, glycoproteins, oligosaccharides, and oligonucleotides.
MALDI is based on the usage of matrix complexed with a given sample molecule that is bombarded with a laser in order for the sample molecule to form a sample ionization. The sample is normally mixed into a high absorbable matrix with as little matrix as possible as the matrix will also become excited and come off and ionize as well. The matrix itself acts as a substance which infuses the sample as well as a transformer for the laser's energy into excitation energy to allow for the vaporization of the sample ions and matrix ions from the surface of the matrix. Most commercially available MALDI mass spectrometers are now a pulsed nitrogen laser of wavelength 337 nm. This method is normally coupled with a Time of Flight detector in order to obtain proper charge-mass ratios and calculate a mass spectrum.

Samples for this procedure are prepared based on retaining homogeneous MALDI sample. This would include focusing on the concentration of the matrix and analyte, the choice of matrix, the analyte sample history, such as how its exposure to strong ionic detergents and formic acid may affect it, also hydrophobicity or hydrophilicity of both the matrix and sample, and possible contaminants and compatible solubilities of matrix and analyte solutions.[4]
SELDI-TOF[edit | edit source]
Surface Enhanced Laser Desorption Ionization is a modification of the procedure used in MALDI. Instead of mixing the UV sensitive matrix with the protein sample, the protein sample is spotted on a plate which has some surface binding characteristics such as a chromatographic array. The spots are then washed to remove impurities and weakly bound proteins. The UV matrix is then added to the spot and allowed to co-crystallize. After the ionization with the UV laser, the ions are analyzed using a TOF mass analyzer, in the same manner as MALDI.
SELDI provides on-chip separation as well as the capability to perform enzymatic reactions directly on the chip. However, there are concerns about the reproducibility of SELDI-TOF mass spectra, especially when normal post processing techniques frequently used with MALDI such as baseline correction are applied. Environmental sources of variation such as humidity can also play a large role.(http://bioinformatics.oxfordjournals.org/cgi/screenpdf/20/5/777 Baggerly et al. 20 (5): 777. (2004))
Tandem MS[edit | edit source]
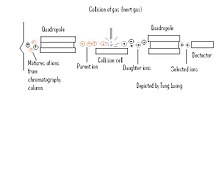
Tandem Mass spectrometry (MS/MS) is spectrometer used to separate ions based on a sample's "electronic" mass using two or more quadrupoles. A sample is normally coupled into tandem MS via chromatography method, normally an electron spray method like gas chromatography or liquid chromatography where the contents are volatilized. More popular tandem mass spectrometers are quadrupole-quadrupole, magnetic sector-quadrupole , and the quadrupole-time-of-flight geometries (P. Roepstorrf, 1989)
The Tandem MS is constructed of two or more quadrupoles, with a collision cell separating each quadrupole. Once a sample has been separated by chromatography, the substances initially go through an initial quadrupole which separates the mixture of ions allotting only certain ions (Pre-cursor ions), passage to the collision cell. The first quadropole is used to select user-specified sample ions from a specific component; usually the molecular-related (i.e (M+H)+ or (M-H)-) ions. Within the collision cell the Precursor-ions also known as "Parent ions" are then bombarded with an inert gas (Xe, Ar, ect ) and are further broken down into different charged and mass ions (Product ions). These product ions also known as "Daughter ions" are then run through an additional quadrupole to further separate the ions which is set to monitor specific ion fragments. This process can be repeated several times in order to get highly specific readings.
In peptide sequencing, selected ion components are either positive (M+H)+ or negative (M-H)- ions. The selected ions will pass into the collision cell, there are three different types of bonds that can fragment along the amino acid backbone: NH-CH, CH-CO, and CO-NH. For each of these fragments, the charge can be maintained by either the N or C-terminal fragment. There are actually six different types of fragments that can be formed. The mass difference between two fragments of the same type is indicative of a particular amino acid. [5]
This technique is primarily used with peptide sequencing due to the parent-daughter ion scanning. Due to the two ions being scanned initially as a precursor and product ion, a mass fingerprinting can occur where through further analysis and investigation the fragments of ions can be pieced together to determine a peptide sequence. Since it is known that amino acid backbone fragments among the NH-CH, CH-CO, and CO-NH bonds, each type of bond can give rise to six different type of species, and these can then be pieced together to provide peptide fragmentation.
TIMS (Thermal Ionization-Mass Spectrometry)[edit | edit source]
TIMS (Thermal Ionization-Mass Spectrometry) is a mass spectrometer that can make exact measurements of isotope ratios of thermally ionizable elements. This ionization can be done by passing them through metal ribbons under vacuum. The resulting ions are accelerated by electrical potential gradient and they are focused into a beam using electrostatically charged plates and slits. The beam is passed through a magnetic field and based on the m/z ratio the original beam produces many separated beams. Using collectors, these separated beams are converted into voltage and exact isotope ratios can be obtained by comparing these beams to their individual ion beams[6].
SSMS (Spark Source Mass Spectrometry)[edit | edit source]
SSMS (Spark Source Mass Spectrometry) can ionize the analytes in solid samples using electric current with two electrodes. It works as one electrode if the sample is metal or can be placed in a cup-shaped electrode by mixing with graphite[7].
Rarely used Mass Spectrometry Ionization Types[edit | edit source]
- Fast Atom Bombardment (FAB): A technique where the anylate is bombarded with highly charged atoms in a vacuum under in a non-volatile matrix.
- Soft laser desorption (SLD): Often referred to as Desorption/Ionization on Silicon (DIOS), is the process by which large macro-molecules are ionized without fragmentation.
- Atmospheric pressure chemical ionization (APCI): A form of chemical ionization unique in that the entire procedure may be performed in atmospheric pressure.
- Secondary ion mass spectrometry (SIMS): A technique which allows for the inspection of solid surfaces via an ion beam disassociation technique called sputtering.
- Spark ionization (SI): Sometimes referred to by the antiquated term spark source ionization, this method allows for the generation of gaseous phase ions from a solid surface.
- Thermal ionization (TI): Often referred to as Surface Ionization, where an anylate is ionzaed from a matrix or filament, usually a metal disk.
References[edit | edit source]
- Coupling Gas Chromatography to Mass Spectrometry, http://www.shsu.edu/~chemistry/primers/gcms.html
- Flash animation of GC-MS, http://www.shsu.edu/~chm_tgc/sounds/sound.html
- How is mass spectrometry used with GC, LC and other separation techniques?, http://www.asms.org/whatisms/p13.html
- Merenbloom, S.I., Koeniger, S.L., Valentine, S.J., Plasencia, M.D., and Clemmer, D.E.; IMS-IMS and IMS-IMS-IMS/MS for Separating Peptide and Protein Fragment Ions; Anal. Chem., 78, 8, 2802 - 2809, 2006
- Sysoev A, Adamov A, Viidanoja J, Ketola RA, Kostiainen R, Kotiaho T. Development of an ion mobility spectrometer for use in an atmospheric pressure ionization ion mobility spectrometer/mass spectrometer instrument for fast screening analysis. 2004 Rapid Commun Mass Spectrom;18(24):3131-9.
- Owens, K. G.; How well do we REALLY understand MALDI?; 4th Annual NIST Polymer MS Workshop, November 9, 2005, Retrieved from http://polymers.msel.nist.gov/maldirecipes/PDF/Owens.pdf
- NCT Proteomics Group - Surface-enhanced laser desorption ionization - time of flight (SELDI-TOF), http://dir.niehs.nih.gov/proteomics/emerg3.htm
- What is mass spectrometry/mass spectrometry (MS/MS)?, http://www.asms.org/whatisms/p15.html
- Understanding the Tandem Mass Spectrometry of Peptides
- ICP-MS, http://web.missouri.edu/~umcreactorweb/pages/ac_icpms1.shtml
- http://en.wikipedia.org/wiki/Accelerator_mass_spectrometry
- Geochemical Instrumentation and Analysis, http://serc.carleton.edu/research_education/geochemsheets/techniques/TIMS.html
- Mass Spectrometry Ionization Methods, http://www.chemistry.adelaide.edu.au/external/soc-rel/content/ionizatn.htm
- http://en.wikipedia.org/wiki/Isotope_ratio_mass_spectrometry

