Principles of Biochemistry/Cell membrane and receptors
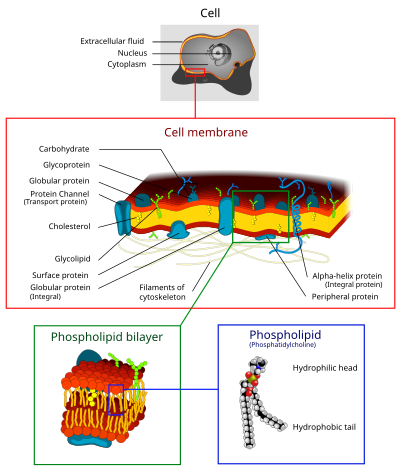
The cell membrane is a biological membrane that separates the interior of all cells from the outside environment. The cell membrane is selectively-permeable to ions and organic molecules and controls the movement of substances in and out of cells. It consists of the phospholipid bilayer with embedded proteins. Cell membranes are involved in a variety of cellular processes such as cell adhesion, ion conductivity and cell signaling and serve as the attachment surface for the extracellular glycocalyx and cell wall and intracellular cytoskeleton.
Composition of Cell membrane[edit | edit source]
Cell membranes contain a variety of biological molecules, notably lipids and proteins. Material is incorporated into the membrane, or deleted from it, by a variety of mechanisms:
- Fusion of intracellular vesicles with the membrane (exocytosis) not only excretes the contents of the vesicle but also incorporates the vesicle membrane's components into the cell membrane. The membrane may form blebs around extracellular material that pinch off to become vesicles (endocytosis).
- If a membrane is continuous with a tubular structure made of membrane material, then material from the tube can be drawn into the membrane continuously.
- Although the concentration of membrane components in the aqueous phase is low (stable membrane components have low solubility in water), there is an exchange of molecules between the lipid and aqueous phases.
Lipids[edit | edit source]
The cell membrane consists of three classes of amphipathic lipids: phospholipids, glycolipids, and cholesterols. The amount of each depends upon the type of cell, but in the majority of cases phospholipids are the most abundant. In RBC studies, 30% of the plasma membrane is lipid.
The fatty chains in phospholipids and glycolipids usually contain an even number of carbon atoms, typically between 16 and 20. The 16- and 18-carbon fatty acids are the most common. Fatty acids may be saturated or unsaturated, with the configuration of the double bonds nearly always cis. The length and the degree of unsaturation of fatty acid chains have a profound effect on membrane fluidity[1] as unsaturated lipids create a kink, preventing the fatty acids from packing together as tightly, thus decreasing the melting temperature (increasing the fluidity) of the membrane. The ability of some organisms to regulate the fluidity of their cell membranes by altering lipid composition is called homeoviscous adaptation.
The entire membrane is held together via non-covalent interaction of hydrophobic tails, however the structure is quite fluid and not fixed rigidly in place. Under physiological conditions phospholipid molecules in the cell membrane are in the liquid crystalline state. It means the lipid molecules are free to diffuse and exhibit rapid lateral diffusion along the layer in which they are present. However, the exchange of phospholipid molecules between intracellular and extracellular leaflets of the bilayer is a very slow process. Lipid rafts and caveolae are examples of cholesterol-enriched microdomains in the cell membrane.
In animal cells cholesterol is normally found dispersed in varying degrees throughout cell membranes, in the irregular spaces between the hydrophobic tails of the membrane lipids, where it confers a stiffening and strengthening effect on the membrane.
Phospholipids forming lipid vesicles[edit | edit source]
Lipid vesicles or liposomes are circular pockets that are enclosed by a lipid bilayer. These structures are used in laboratories to study the effects of chemicals in cells by delivering these chemicals directly to the cell, as well as getting more insight into cell membrane permeability. Lipid vesicles and liposomes are formed by first suspending a lipid in an aqueous solution then agitating the mixture through sonication, resulting in a uniformly circular vesicle. By measuring the rate of efflux from that of the insideof the vesicle to the ambient solution, allows researcher to better understand membrane permeability. Vesicles can be formed with molecules and ions inside the vesicle by forming the vesicle with the desired molecule or ion present in the solution. Proteins can also be embedded into the membrane through solubilizing the desired proteins in the presence of detergents and attaching them to the phospholipids in which the liposome is formed. These provide researchers with a tool to examine various membrane protein functions.
What is a Vesicle?[edit | edit source]
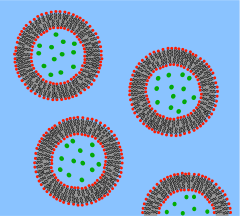
A vesicle is a lipid bilayer rolled up into a spherical shell, enclosing a small amount of water and separating it from the water outside the vesicle. Because of this fundamental similarity to the cell membrane, vesicles have been used extensively to study the properties of lipid bilayers. Another reason vesicles have been used so frequently is that they are relatively easy to make. If a sample of dehydrated lipid is exposed to water it will spontaneously form vesicles.[2] These initial vesicles are typically multilamellar (many-walled) and are of a wide range of sizes from tens of nanometers to several micrometres.[3] Methods such as sonication or extrusion through a membrane are needed to break these initial vesicles into smaller, single-walled vesicles of uniform diameter known as small unilamellar vesicles (SUVs). SUVs are typically between 50 and 200 nm diameter.[4] Alternatively, rather than synthesizing vesicles it is possible to simply isolate them from cell cultures or tissue samples.[5] Vesicles are used to transport lipids, proteins and many other molecules within the cell as well as into or out of the cell. These naturally isolated vesicles are composed of a complex mixture of different lipids and proteins so, although they offer greater realism for studying specific biological phenomena, simple artificial vesicles are preferred for studies of fundamental lipid properties.
Since artificial SUVs can be made in large quantities they are suitable for bulk material studies such as x-ray diffraction to determine lattice spacing[6] and differential scanning calorimetry to determine phase transitions.[7] Dual polarisation interferometry can measure unilamelar and multilamelar structures and insertion into and disruption of the vesciles in a label free assay format [8]. Vesicles can also be labeled with fluorescent dyes to allow sensitive FRET-based fusion assays.[9] In spite of this fluorescent labeling it is often difficult to perform detailed imaging on SUVs simply because they are so small. To combat this problem researchers have developed the giant unilamellar vesicle (GUV). GUVs are large enough (several tens of micrometres) to study with traditional florescence microscopy. Many of the studies of lipid rafts in artificial lipid systems have been performed with GUVs for this reason.[10] Compared to supported bilayers, GUVs present a more “natural” environment since there is no nearby solid surface to induce defects or denature proteins. However, GUVs are relatively fragile, time consuming to make and can only be produced in limited yield compared to SUVs.
To circumvent these problems a microfluidic assembly line approach to GUVs was reported.[11]
Carbohydrates[edit | edit source]
Plasma membranes also contain carbohydrates, predominantly glycoproteins, but with some glycolipids (cerebrosides and gangliosides). For the most part, no glycosylation occurs on membranes within the cell; rather generally glycosylation occurs on the extracellular surface of the plasma membrane.
The glycocalyx is an important feature in all cells, especially epithelia with microvilli. Recent data suggest the glycocalyx participates in cell adhesion, lymphocyte homing, and many others.
The penultimate sugar is galactose and the terminal sugar is sialic acid, as the sugar backbone is modified in the golgi apparatus. Sialic acid carries a negative charge, providing an external barrier to charged particles.
Proteins[edit | edit source]
Proteins within the membrane are key to the functioning of the overall membrane. These proteins mainly transport chemicals and information across the membrane. Every membrane has a varying degree of protein content. Proteins can be in the form of peripheral or integral.
| Type | Description | Examples |
| Integral proteins or transmembrane proteins |
Span the membrane and have a hydrophilic cytosolic domain, which interacts with internal molecules, a hydrophobic membrane-spanning domain that anchors it within the cell membrane, and a hydrophilic extracellular domain that interacts with external molecules. The hydrophobic domain consists of one, multiple, or a combination of α-helices and β sheet protein motifs. | Ion channels, proton pumps, G protein-coupled receptor |
| Lipid anchored proteins | Covalently-bound to single or multiple lipid molecules; hydrophobically insert into the cell membrane and anchor the protein. The protein itself is not in contact with the membrane. | G proteins |
| Peripheral proteins | Attached to integral membrane proteins, or associated with peripheral regions of the lipid bilayer. These proteins tend to have only temporary interactions with biological membranes, and, once reacted the molecule, dissociates to carry on its work in the cytoplasm. | Some enzymes, some hormones |
The cell membrane plays host to a large amount of protein that is responsible for its various activities. The amount of protein differs between species and according to function, however the typical amount in a cell membrane is 50%.[1] These proteins are undoubtedly important to a cell: Approximately a third of the genes in yeast code specifically for them, and this number is even higher in multicellular organisms.
The cell membrane, being exposed to the outside environment, is an important site of cell-cell communication. As such, a large variety of protein receptors and identification proteins, such as antigens, are present on the surface of the membrane. Functions of membrane proteins can also include cell-cell contact, surface recognition, cytoskeleton contact, signaling, enzymatic activity, or transporting substances across the membrane.
Most membrane proteins must be inserted in some way into the membrane. For this to occur, an N-terminus "signal sequence" of amino acids directs proteins to the endoplasmic reticulum, which inserts the proteins into a lipid bilayer. Once inserted, the proteins are then transported to their final destination in vesicles, where the vesicle fuses with the target membrane.

The membrane is represented in light brown.
Transmembrane protein (TP)[edit | edit source]
A transmembrane protein (TP) is a protein that goes from one side of a membrane through to the other side of the membrane. Many TPs function as gateways or "loading docks" to deny or permit the transport of specific substances across the biological membrane, to get into the cell, or out of the cell as in the case of waste byproducts. As a response to the shape of a cerain molecules these "freight handling" TPs may have special ways of folding up or bending that will move a substance through the biological membrane. A transmembrane protein is a polytopic protein that spans an entire biological membrane. Transmembrane proteins aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
All transmembrane proteins are integral membrane proteins, but not all IMPs are transmembrane proteins.
There are two basic types of transmembrane proteins: Alpha-helical. These proteins are present in the inner membranes of bacterial cells or the plasma membrane of eukaryotes, and sometimes in the outer membranes. This is the major category of transmembrane proteins. In humans, 27% of all proteins have been estimated to be alpha-helical membrane proteins. Beta-barrels. These proteins are so far found only in outer membranes of Gram-negative bacteria, cell wall of Gram-positive bacteria, and outer membranes of mitochondria and chloroplasts. All beta-barrel transmembrane proteins have simplest up-and-down topology, which may reflect their common evolutionary origin and similar folding mechanism. Another classification refers to the position of the N- and C-terminal domains. Types I, II, and III are single pass molecules, while type IV are multiple pass molecules. Type I transmembrane proteins are anchored to the lipid membrane with a stop-transfer anchor sequence and have their N-terminal domains targeted to the ER lumen during synthesis (and the extracellular space, if mature forms are located on plasmalemma). Type II and III are anchored with a signal-anchor sequence, with type II being targeted to the ER lumen with its C-terminal domain, while type III have their N-terminal domains targeted to the ER lumen. Type IV is subdivided into IV-A, with their N-terminal domains targeted to the cytosol and IV-B, with an N-terminal domain targeted to the lumen. The implications for the division in the four types are especially manifest at the time of translocation and ER-bound translation, when the protein has to be passed through the ER membrane in a direction dependent on the type.
Integral membrane protein (IMP)[edit | edit source]
integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins. An integral membrane protein (IMP) is a protein molecule (or assembly of proteins) that is permanently attached to the biological membrane. Proteins that cross the membrane are surrounded by "annular" lipids, which are defined as lipids that are in direct contact with a membrane protein. Such proteins can be separated from the biological membranes only using detergents, nonpolar solvents, or sometimes denaturing agents. IMPs comprise a very significant fraction of the proteins encoded in an organism's genome. All transmembrane proteins are IMPs, but not all IMPs are transmembrane proteins.
Peripheral membrane proteins[edit | edit source]
Peripheral membrane proteins are proteins that adhere only temporarily to the biological membrane with which they are associated. These molecules attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins. The reversible attachment of proteins to biological membranes has shown to regulate cell signaling and many other important cellular events, through a variety of mechanisms.For example, the close association between many enzymes and biological membranes may bring them into close proximity with their lipid substrate(s). Membrane binding may also promote rearrangement, dissociation, or conformational changes within many protein structural domains, resulting in an activation of their biological activity. Additionally, the positioning of many proteins are localized to either the inner or outer surfaces or leaflets of their resident membrane. This facilitates the assembly of multi-protein complexes by increasing the probability of any appropriate protein-protein interactions.
Peripheral membrane proteins may interact with other proteins or directly with the lipid bilayer. In the latter case, they are then known as amphitropic proteins. Some proteins, such as G-proteins and certain protein kinases, interact with transmembrane proteins and the lipid bilayer simultaneously. Some polypeptide hormones, antimicrobial peptides, and neurotoxins accumulate at the membrane surface prior to locating and interacting with their cell surface receptor targets, which may themselves be peripheral membrane proteins. The Phospholipid bilayer that forms the cell surface membrane consists of a hydrophobic inner core region sandwiched between two regions of hydrophilicity, one at the inner surface and one at the outer surface of the cell membrane (see lipid bilayer article for a more detailed structural description of the cell membrane). The inner and outer surfaces, or interfacial regions, of model phospholipid bilayers have been shown to have a thickness of around 8 to 10 Å, although this may be wider in biological membranes that include large amounts of gangliosides or lipopolysaccharides. The hydrophobic inner core region of typical biological membranes may have a thickness of around 27 to 32 Å, as estimated by Small angle X-ray scattering (SAXS). The boundary region between the hydrophobic inner core and the hydrophilic interfacial regions is very narrow, at around 3Å, (see lipid bilayer article for a description of its component chemical groups). Moving outwards away from the hydrophobic core region and into the interfacial hydrophilic region, the effective concentration of water rapidly changes across this boundary layer, from nearly zero to a concentration of around 2M.[8][9] The phosphate groups within phospholipid bilayers are fully hydrated or saturated with water and are situated around 5 Å outside the boundary of the hydrophobic core region (see Figures ). Some water-soluble proteins associate with lipid bilayers irreversibly and can form transmembrane alpha-helical or beta-barrel channels. Such transformations occur in pore forming toxins such as colicin A, alpha-hemolysin, and others. They may also occur in [[Bcl-2-associated X protein|BcL-2 like protein , in some amphiphilic antimicrobial peptides , and in certain annexins . These proteins are usually described as peripheral as one of their conformational states is water-soluble or only loosely associated with a membrane.
Receptor[edit | edit source]
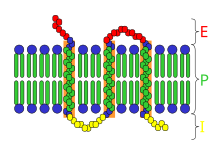
In biochemistry, a receptor is a protein molecule, embedded in either the plasma membrane or the cytoplasm of a cell, to which one or more specific kinds of signaling molecules may attach. A molecule which binds (attaches) to a receptor is called a ligand, and may be a peptide (short protein) or other small molecule, such as a neurotransmitter, a hormone, a pharmaceutical drug, or a toxin. Each kind of receptor can bind only certain ligand shapes. Each cell typically has many receptors, of many different kinds. Simply put, a receptor functions as a keyhole that opens a neural path when the proper ligand is inserted. Ligand binding stabilizes a certain receptor conformation (the three-dimensional shape of the receptor protein, with no change in sequence). This is often associated with gain of or loss of protein activity, ordinarily leading to some sort of cellular response. However, some ligands (e.g. antagonists) merely block receptors without inducing any response. Ligand-induced changes in receptors result in cellular changes which constitute the biological activity of the ligands. Many functions of the human body are regulated by these receptors responding uniquely to specific molecules like this.
The shapes and actions of receptors are studied by X-ray crystallography, dual polarisation interferometry, computer modelling, and structure-function studies, which have advanced the understanding of drug action at the binding sites of receptors. Structure activity relationships correlate induced conformational changes with biomolecular activity, and are studied using dynamic techniques such as circular dichroism and dual polarisation interferometry.
Depending on their functions and ligands, several types of receptors may be identified: Some receptor proteins are peripheral membrane proteins. Many hormone and neurotransmitter receptors are transmembrane proteins: transmembrane receptors are embedded in the phospholipid bilayer of cell membranes, that allow the activation of signal transduction pathways in response to the activation by the binding molecule, or ligand. Metabotropic receptors are coupled to G proteins and affect the cell indirectly through enzymes which control ion channels. Ionotropic receptors (also known as ligand-gated ion channels) contain a central pore which opens in response to the binding of ligand. Another major class of receptors are intracellular proteins such as those for steroid and intracrine peptide hormone receptors. These receptors often can enter the cell nucleus and modulate gene expression in response to the activation by the ligand. Membrane receptors are isolated from cell membranes by complex extraction procedures using solvents, detergents, and/or affinity purification.
Metabotropic receptor[edit | edit source]
Metabotropic receptor is a subtype of membrane receptors at the surface or in vesicles of eukaryotic cells. In the nervous system, based on their structural and functional characteristics, neurotransmitter receptors can be classified into two broad categories: metabotropic and ionotropic receptors. In contrast to the latter, metabotropic receptors do not form an ion channel pore; rather, they are indirectly linked with ion-channels on the plasma membrane of the cell through signal transduction mechanisms, often G proteins. Hence, they are a type of G protein-coupled receptor. Others are tyrosine kinases or guanylyl cyclase receptors. What both receptor types have in common is that they are activated by specific neurotransmitters. When an ionotropic receptor is activated, it opens a channel that allows ions such as Na+, K+, or Cl- to flow. In contrast, when a metabotropic receptor is activated, a series of intracellular events are triggered that also results in ion channel opening but must involve a range of second messenger chemicals.
Ligand-gated ion channels (LGICs)[edit | edit source]
Ligand-gated ion channels (LGICs) are one type of ionotropic receptor or channel-linked receptor. They are a group of transmembrane ion channels that are opened or closed in response to the binding of a chemical messenger (i.e., a ligand), such as a neurotransmitter. The binding site of endogenous ligands on LGICs protein complexes are normally located on a different portion of the protein (an allosteric binding site) compared to where the ion conduction pore is located. The direct link between ligand binding and opening or closing of the ion channel, which is characteristic of ligand-gated ion channels, is contrasted with the indirect function of metabotropic receptors, which use second messengers. LGICs are also different from voltage-gated ion channels (which open and close depending on membrane potential), and stretch-activated ion channels (which open and close depending on mechanical deformation of the cell membrane).
G protein-coupled receptors (GPCRs)[edit | edit source]
G protein-coupled receptors (GPCRs), also known as seven-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptor, and G protein-linked receptors (GPLR), comprise a large protein family of transmembrane receptors that sense molecules outside the cell and activate inside signal transduction pathways and, ultimately, cellular responses. G protein-coupled receptors are found only in eukaryotes, including yeast, choanoflagellates, and animals. The ligands that bind and activate these receptors include light-sensitive compounds, odors, pheromones, hormones, and neurotransmitters, and vary in size from small molecules to peptides to large proteins. G protein-coupled receptors are involved in many diseases, and are also the target of approximately 30% of all modern medicinal drugs.
The exact size of the GPCR superfamily is unknown but nearly 800 different human genes (or ≈4% of the entire protein-coding genome) have been predicted from genome sequence analysis. Although numerous classification schemes have been proposed, the superfamily is classically divided into three main classes (A, B, and C) with no detectable shared sequence homology between classes. The largest class by far is class A, which accounts for nearly 85% of the GPCR genes. Of class A GPCRs, over half of these are predicted to encode olfactory receptors while the remaining receptors are liganded by known endogenous compounds or are classified as orphan receptors. Despite the lack of sequence homology between classes, all GPCRs share a common structure and mechanism of signal transduction.

In all, GPCRs can be grouped into 6 classes based on sequence homology and functional similarity:
Class A (or 1) (Rhodopsin-like)
Class B (or 2) (Secretin receptor family)
Class C (or 3) (Metabotropic glutamate/pheromone)
Class D (or 4) (Fungal mating pheromone receptors)
Class E (or 5) (Cyclic AMP receptors)
Class F (or 6) (Frizzled/Smoothened)
The very large rhodopsin A group has been further subdivided into 19 subgroups (A1-A19). More recently, an alternative classification system called GRAFS (Glutamate, Rhodopsin, Adhesion, Frizzled/Taste2, Secretin) has been proposed. The human genome encodes thousands of G protein-coupled receptors, about 350 of which detect hormones, growth factors, and other endogenous ligands. Approximately 150 of the GPCRs found in the human genome have unknown functions. Some web-servers and bioinformatics prediction methods have been used for predicting the classification of GPCRs according to their amino acid sequence alone, by means of the pseudo amino acid composition approach.
Structure of GPCRs[edit | edit source]
Structurally GPCRs are characterized by an extracellular N-terminus, followed by seven transmembrane (7-TM) α-helices (TM-1 to TM-7) connected by three intracellular (IL-1 to IL-3) and three extracellular loops (EL-1 to EL-3), and finally an intracellular C-terminus. The GPCR arranges itself into a tertiary structure resembling a barrel, with the seven transmembrane helices forming a cavity within the plasma membrane which serves a ligand-binding domain that is often covered by EL-2. Ligands may also bind elsewhere, however, as is the case for bulkier ligands (e.g., proteins or large peptides) which instead interact with the extracellular loops, or, as illustrated by the class C metabotropic glutamate receptors (mGluRs), the N-terminal tail. The class C GPCRs are distinguished by their large N-terminal tail, which also contains a ligand-binding domain. Upon glutamate-binding to an mGluR, the N-terminal tail undergoes a conformational change that leads to its interaction with the residues of the extracellular loops and TM domains. The eventual effect of all three types of agonist-induced activation is a change in the relative orientations of the TM helices (likened to a twisting motion) leading to a wider intracellular surface and “revelation” of residues of the intracellular helices and TM domains crucial to signal transduction function (i.e., G-protein coupling). Inverse agonists and antagonists may also bind to a number of different sites, but the eventual effect must be prevention of this TM helix reorientation. The structure of the N- and C-terminal tails of GPCRs may also serve important functions beyond ligand-binding. In particular, the C-terminus often contains serine (Ser) or threonine (Thr) residues that, when phosphorylated, increase the affinity of the intracellular surface for the binding of scaffolding proteins called β-arrestins (β-arr). Once bound, β-arrestins both sterically prevent G-protein coupling and may recruit other proteins leading to the creation of signaling complexes involved in extracellular-signal regulated kinase (ERK) pathway activation or receptor endocytosis (internalization). As the phosphorylation of these Ser and Thr residues often occurs as a result of GPCR activation, the β-arr-mediated G-protein-decoupling and internalization of GPCRs are important mechanisms of desensitization. A final common structural theme amongst GPCRs is palmitoylation of one or more sites of the C-terminal tail or the intracellular loops. Palmitoylation is the covalent modification of cysteine (Cys) residues via addition of hydrophobic acyl groups, and has the effect of targeting the receptor to cholesterol- and sphingolipid-rich microdomains of the plasma membrane called lipid rafts. As many of the downstream transducer and effector molecules of GPCRs (including those involved in negative feedback pathways) are also targeted to lipid rafts, this has the effect of facilitating rapid receptor signaling. GPCRs respond to extracellular signals mediated by a huge diversity of agonists, ranging from proteins to biogenic amines to protons, but all transduce this signal via a mechanism of G-protein coupling. This is made possible by virtue of a guanine-nucleotide exchange factor (GEF) domain primarily formed by a combination of IL-2 and IL-3 along with adjacent residues of the associated TM helices.
Receptor tyrosine kinases (RTK)s[edit | edit source]
Receptor tyrosine kinases (RTK)s are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. Of the 90 unique tyrosine kinase genes identified in the human genome, 58 encode receptor tyrosine kinase proteins. Receptor tyrosine kinases have been shown not only to be key regulators of normal cellular processes but also to have a critical role in the development and progression of many types of cancer.
Most RTKs are single subunit receptors but some exist as multimeric complexes, e.g., the insulin receptor that forms disulfide-linked dimers in the absence of hormone; moreover, ligand binding to the extracellular domain induces formation of receptor dimers. Each monomer has a single hydrophobic transmembrane-spanning domain composed of 25-38 amino acids, an extracellular N-terminal region, and an intracellular C-terminal region. The extracellular N-terminal region exhibits a variety of conserved elements including immunoglobulin (Ig)-like or epidermal growth factor (EGF)-like domains, fibronectin type III repeats, or cysteine-rich regions that are characteristic for each subfamily of RTKs; these domains contain primarily a ligand-binding site, which binds extracellular ligands, e.g., a particular growth factor or hormone. The intracellular C-terminal region displays the highest level of conservation and comprises catalytic domains responsible for the kinase activity of these receptors, which catalyses receptor autophosphorylation and tyrosine phosphorylation of RTK substrates.
Families of RTKs[edit | edit source]
Epidermal growth factor receptor family
The ErbB protein family or epidermal growth factor receptor (EGFR) family is a family of four structurally related receptor tyrosine kinases. Insufficient ErbB signaling in humans is associated with the development of neurodegenerative diseases, such as multiple sclerosis and Alzheimer's Disease.In mice, loss of signaling by any member of the ErbB family results in embryonic lethality with defects in organs including the lungs, skin, heart, and brain. Excessive ErbB signaling is associated with the development of a wide variety of types of solid tumor. ErbB-1 and ErbB-2 are found in many human cancers and their excessive signaling may be critical factors in the development and malignancy of these tumors.
Fibroblast growth factor receptor (FGFR) family
Fibroblast growth factors comprise the largest family of growth factor ligands at 23 members. The natural alternate splicing of four fibroblast growth factor receptor (FGFR) genes results in the production of over 48 different isoforms of FGFR.These isoforms vary in their ligand binding properties and kinase domains; however, all share a common extracellular region composed of three immunoglobulin (Ig) like domains (D1-D3), and thus belong to the immunoglobulin superfamily. Interactions with FGFs occur via FGFR domains D2 and D3. Each receptor can be activated by several FGFs. In many cases the FGFs themselves can also activate more than one receptor, this is not the case with FGF-7, however, which can activate only FGFR2b. A gene for a fifth FGFR protein, FGFR5, has also been identified. In contrast to FGFRs 1-4 it lacks a cytoplasmic tyrosine kinase domain and one isoform, FGFR5γ, only contains the extracellular domains D1 and D2.
Vascular endothelial growth factor receptor (VEGFR) family
Vascular endothelial growth factor (VEGF) is one of the main inducers of endothelial cell proliferation and permeability of blood vessels. Two RTKs bind to VEGF at the cell surface, VEGFR-1 (Flt-1) and VEGFR-2 (KDR/Flk-1). The VEGF receptors have an extracellular portion consisting of seven Ig-like domains so, like FGFRs, belong to the immunoglobulin superfamily. They also possess a single transmembrane spanning region and an intracellular portion containing a split tyrosine-kinase domain. VEGF-A binds to VEGFR-1 (Flt-1) and VEGFR-2 (KDR/Flk-1). VEGFR-2 appears to mediate almost all of the known cellular responses to VEGF. The function of VEGFR-1 is less well defined, although it is thought to modulate VEGFR-2 signaling. Another function of VEGFR-1 may be to act as a dummy/decoy receptor, sequestering VEGF from VEGFR-2 binding (this appears to be particularly important during vasculogenesis in the embryo). A third receptor has been discovered (VEGFR-3); however, VEGF-A is not a ligand for this receptor. VEGFR-3 mediates lymphangiogenesis in response to VEGF-C and VEGF-D.
RET receptor family
The natural alternate splicing of the RET gene results in the production of 3 different isoforms of the protein RET. RET51, RET43, and RET9 contain 51, 43, and 9 amino acids in their C-terminal tail, respectively. The biological roles of isoforms RET51 and RET9 are the most well studied in-vivo as these are the most common isoforms in which RET occurs. RET is the receptor for members of the glial cell line-derived neurotrophic factor (GDNF) family of extracellular signalling molecules or ligands (GFLs). In order to activate RET, first GFLs must form a complex with a glycosylphosphatidylinositol (GPI)-anchored co-receptor. The co-receptors themselves are classified as members of the GDNF receptor-α (GFRα) protein family. Different members of the GFRα family (GFRα1-GFRα4) exhibit a specific binding activity for a specific GFLs. Upon GFL-GFRα complex formation, the complex then brings together two molecules of RET, triggering trans-autophosphorylation of specific tyrosine residues within the tyrosine kinase domain of each RET molecule. Phosphorylation of these tyrosines then initiates intracellular signal transduction processes.
Eph receptor Family
Ephrin and Eph receptors are the largest subfamily of RTKs.
References[edit | edit source]
- ↑ a b Jesse Gray, Shana Groeschler, Tony Le, Zara Gonzalez (2002). "Membrane Structure" (SWF). Davidson College. Retrieved 2007-01-11.
{{cite web}}: CS1 maint: multiple names: authors list (link) - ↑ A D Bangham and R W Horne."Negative stainign of phospholipids and their structural modification by surface active agents as observed in the electron microscope." Journal of Molecular Biology. 8. (1964) 660-668.
- ↑ D D Lasic."The mechanism of vesicle formation." Biochemical Journal. 256. (1988) 1-11.
- ↑ F Szoka and D Papahadjopoulos."Comparative Properties and Methods of Preparation of Lipid Vesicles (Liposomes)." Annual Review of Biophysics and Bioengineering. 9. (1980) 467-508.
- ↑ W S Trimble, D M Cowan, and R H Scheller."VAMP-1: a synaptic vesicle-associated integral membrane protein." Proceedings of the National Academy of Sciences of the United States of America. 85. (1988) 4538-4542.
- ↑ D Papahadjapoulos and N Miller."Phospholipid Model Membranes I. Structural characteristics of hydrated liquid crystals." Biochimica et Biophysica Acta. 135. (1967) 624-638.
- ↑ H Trauble and D H Haynes."The volume change in lipid bilayer lamellae at the crystalline-liquid crystalline phase transition." Chem. Phys. Lipids. 7. (1971) 324-335.
- ↑ J Popplewell, M Swann, N Freeman, C McDonnell and R Ford, "Quantifying of the Effects of Mellitin on Liposomes." Biochimica et Biophysica Acta (2007) 1768 13-20
- ↑ L Guohua and R C Macdonald."Lipid bilayer vesicle fusion: Intermediates captured by high-speed microfluorescence spectroscopy." Biophysical Journal. 85. (2003) 1585-1599.
- ↑ C Dietrich, L A Bagatolli, Z N Volovyk, N L Thompson, et al."Lipid rafts reconstituted in model membranes." Biophysical Journal. 80. (2001) 1417-1428.
- ↑ Matosevic, S.; Paegel, B. J. Am. Chem. Soc. Article ASAP DO1: 10.1021/ja109137s